User:RotogenRay/Receptors
Receptors and their associated ligands, agonists, antagonists, second messengers, etc. play an important role in facilitating all organisms' ability to respond to the environment. Likewise, categorization and contectualization of receptor families is tantamount to elucidate understanding of their function in the organisim. However, the challenge with categorization and current efforts to that end is determining a common denominator, or criteria for classification. With such a wide variety of known receptors and associated genetics, and many still unknown and currently researched cellular sensory and signal transduction mechanisms, and given the fact that different parts of the body tend to use certain receptor systems in different ways, there is a disconnect between the observation of receptor structure, location-specific function, and the larger effect on the whole organism to the presence of metabolic response modifiers, an excess or defficiet of ligands, receptor agonists, antagonists, cofactors and second messengers, etc.
Genetic Classification
[edit]Ion channel genes
[edit](from Ion_channel_family
Ion channels and their associated receptors are most useful when fast neurotransmission or signal transduction is necessary. However many ion channels serve regulation or passive functions.
Human channels with six transmembrane helices in each subunit
[edit]Cation
- MCOLN1; MCOLN2; MCOLN3;
- PKD1L3;
- TRPA1
- TPCN1; TPCN2
- TRPC1: TRPC3; TRPC4; TRPC5; TRPC6; TRPC7
- TRPM1; TRPM2; TRPM3; TRPM4; TRPM5; TRPM6; TRPM7; TRPM8
- TRPV1; TRPV2; TRPV3; TRPV4; TRPV5; TRPV6
Calcium
- CACNA1A; CACNA1B; CACNA1C; CACNA1D; CACNA1E; CACNA1F; CACNA1G; CACNA1H; CACNA1I; CACNA1S
- CATSPER1; CATSPER2; CATSPER3; CATSPER4
- RYR1; RYR2; RYR3
Potassium[1]
- KCNA1; KCNA2; KCNA3; KCNA4; KCNA5; KCNA6; KCNA7; KCNA10
- KCNB1; KCNB2
- KCNC1; KCNC2; KCNC3; KCNC4
- KCND1; KCND2; KCND3
- KCNF1
- KCNG1; KCNG2; KCNG3; KCNG4
- KCNH1; KCNH2; KCNH3; KCNH4; KCNH5; KCNH6; KCNH7; KCNH8
- KCNMA1
- KCNQ1; KCNQ2; KCNQ3; KCNQ4; KCNQ5
- KCNS1; KCNS2; KCNS3
- KCNV1; KCNV2
Sodium
- NALCN
- SCN1A; SCN2A; SCN2A2; SCN3A; SCN4A; SCN5A; SCN7A; SCN8A; SCN9A; SCN10A; SCN11A
- SLC9A10; SLC9A11
Proton
Cyclic nucleotide-gated
Human channels with two TM helices in each subunit, as in bacteria
[edit]Potassium
- KCNK1; KCNK2; KCNK3; KCNK4; KCNK5; KCNK6; KCNK7; KCNK9; KCNK10; KCNK12; KCNK13; KCNK15; KCNK16; KCNK17; KCNK18
- KCNN1; KCNN2; KCNN3; KCNN4;
- KCNT1; KCNT2
G coupled protien receptors
[edit]Rhodopsin-like GCPRs
[edit]Subfamily A1
[edit]- Chemokine receptor InterPro: IPR000355
- Chemokine (C-C motif) receptor 1 (CCR1, CKR1)
- Chemokine (C-C motif) receptor 2 (CCR2, CKR2)
- Chemokine (C-C motif) receptor 3 (CCR3, CKR3)
- Chemokine (C-C motif) receptor 4 (CCR4, CKR4)
- Chemokine (C-C motif) receptor 5 (CCR5, CKR5)
- Chemokine (C-C motif) receptor 8 (CCR8, CKR8)
- Chemokine (C-C motif) receptor-like 2 (CCRL2, CKRX)
- chemokine (C motif) receptor 1 (XCR1, CXC1) InterPro: IPR005393
- chemokine (C-X3-C motif) receptor 1 (CX3CR1, C3X1) InterPro: IPR005387
- GPR137B (GPR137B, TM7SF1)
Subfamily A2
[edit]- Chemokine receptor InterPro: IPR000355
- Chemokine (C-C motif) receptor-like 1 (CCRL1 CCRL1, CCR11)
- Chemokine (C-C motif) receptor 6 (CCR6, CKR6)
- Chemokine (C-C motif) receptor 7 (CCR7, CKR7)
- Chemokine (C-C motif) receptor 9 (CCR9, CKR9)
- Chemokine (C-C motif) receptor 10 (CCR10, CKRA)
- CXC chemokine receptors InterPro: IPR001053
- Interleukin-8 InterPro: IPR000174 (IL8R)
- Adrenomedullin receptor (GPR182)
- Duffy blood group, chemokine receptor (DARC, DUFF)
- G Protein-coupled Receptor 30 (GPER, CML2, GPCR estrogen receptor)
Subfamily A3
[edit]- Angiotensin II receptor InterPro: IPR000248
- Angiotensin II receptor, type 1 (AGTR1, AG2S)
- Angiotensin II receptor, type 2 (AGTR2, AG22)
- Apelin receptor (AGTRL1, APJ) InterPro: IPR003904
- Bradykinin receptor InterPro: IPR000496
- Bradykinin receptor B1 (BDKRB1, BRB1)
- Bradykinin receptor B2 (BDKRB2, BRB2)
- GPR15 (GPR15, GPRF)
- GPR25 (GPR25)
Subfamily A4
[edit]- Opioid receptor InterPro: IPR001418
- delta Opioid receptor (OPRD1, OPRD)
- kappa Opioid receptor (OPRK1, OPRK)
- mu Opioid receptor (OPRM1, OPRM)
- Nociceptin receptor (OPRL1, OPRX)
- Somatostatin receptor InterPro: IPR000586
- Somatostatin receptor 1 (SSTR1, SSR1)
- Somatostatin receptor 2 (SSTR2, SSR2)
- Somatostatin receptor 3 (SSTR3, SSR3)
- Somatostatin receptor 4 (SSTR4, SSR4)
- Somatostatin receptor 5 (SSTR5, SSR5)
- GPCR neuropeptide receptor InterPro: IPR009150
- Neuropeptides B/W receptor 1 (NPBWR1, GPR7)
- Neuropeptides B/W receptor 2 (NPBWR2, GPR8)
- GPR1 orphan receptor (GPR1) InterPro: IPR002275
Subfamily A5
[edit]- Galanin receptor InterPro: IPR000405
- Galanin receptor 1 (GALR1, GALR)
- Galanin receptor 2 (GALR2, GALS)
- Galanin receptor 3 (GALR3, GALT)
- Cysteinyl leukotriene receptor InterPro: IPR004071
- Leukotriene B4 receptor InterPro: IPR003981
- Relaxin receptor InterPro: IPR008112
- Relaxin/insulin-like family peptide receptor 1 (RXFP1, LGR7)
- Relaxin/insulin-like family peptide receptor 2 (RXFP2, GPR106)
- Relaxin/insulin-like family peptide receptor 3 (RXFP3, SALPR)
- Relaxin/insulin-like family peptide receptor 4 (RXFP4, GPR100/GPR142)
- KiSS1-derived peptide receptor (GPR54) (KISS1R) InterPro: IPR008103
- Melanin-concentrating hormone receptor 1 (MCHR1, GPRO) InterPro: IPR008361
- Urotensin-II receptor (UTS2R, UR2R) InterPro: IPR000670
Subfamily A6
[edit]- Cholecystokinin receptor InterPro: IPR009126
- Cholecystokinin A receptor (CCKAR, CCKR)
- Cholecystokinin B receptor (CCKBR, GASR)
- Neuropeptide FF receptor InterPro: IPR005395
- Neuropeptide FF receptor 1 (NPFFR1, FF1R)
- Neuropeptide FF receptor 2 (NPFFR2, FF2R)
- Orexin receptor InterPro: IPR000204
- Hypocretin (orexin) receptor 1 (HCRTR1, OX1R)
- Hypocretin (orexin) receptor 2 (HCRTR2, OX2R)
- Vasopressin receptor InterPro: IPR001817
- Arginine vasopressin receptor 1A (AVPR1A, V1AR)
- Arginine vasopressin receptor 1B (AVPR1B, V1BR)
- Arginine vasopressin receptor 2 (AVPR2, V2R)
- Gonadotrophin releasing hormone receptor (GNRHR, GRHR) InterPro: IPR001658
- Pyroglutamylated RFamide peptide receptor (QRFPR, GPR103)
- GPR22 (GPR22, GPRM)
- GPR176 (GPR176, GPR)
Subfamily A7
[edit]- Bombesin receptor InterPro: IPR001556
- Endothelin receptor InterPro: IPR000499
- Endothelin receptor type A (EDNRA, ET1R)
- Endothelin receptor type B (EDNRB, ETBR)
- GPR37 (GPR37, ETBR-LP2) InterPro: IPR003909
- Neuromedin U receptor InterPro: IPR005390
- Neurotensin receptor InterPro: IPR003984
- Neurotensin receptor 1 (NTSR1, NTR1)
- Neurotensin receptor 2 (NTSR2, NTR2)
- Thyrotropin-releasing hormone receptor (TRHR, TRFR) InterPro: IPR009144
- Growth hormone secretagogue receptor (GHSR) InterPro: IPR003905
- GPR39 (GPR39)
- Motilin receptor (MLNR, GPR38)
Subfamily A8
[edit]- Anaphylatoxin receptors InterPro: IPR002234
- C3a receptor (C3AR1, C3AR)
- C5a receptor (C5AR1, C5AR)
- Chemokine-like receptor 1 (CMKLR1, CML1) InterPro: IPR002258
- Formyl peptide receptor InterPro: IPR000826
- Formyl peptide receptor 1 (FPR1, FMLR)
- Formyl peptide receptor-like 1 (FPRL1, FML2)
- Formyl peptide receptor-like 2 (FPRL2, FML1)
- MAS1 oncogene InterPro: IPR000820
- GPR1 (GPR1)
- GPR32 (GPR32, GPRW)
- GPR44 (GPR44)
- GPR77 (GPR77, C5L2)
Subfamily A9
[edit]- Melatonin receptor InterPro: IPR000025
- Melatonin receptor 1A (MTNR1A, ML1A)
- Melatonin receptor 1B (MTNR1B, ML1B)
- Neurokinin receptor InterPro: IPR001681
- Tachykinin receptor 1 (TACR1, NK1R)
- Tachykinin receptor 2 (TACR2, NK2R)
- Tachykinin receptor 3 (TACR3, NK3R)
- Neuropeptide Y receptor InterPro: IPR000611
- Neuropeptide Y receptor Y1 (NPY1R, NY1R)
- Neuropeptide Y receptor Y2 (NPY2R, NY2R)
- Pancreatic polypeptide receptor 1 (PPYR1, NY4R)
- Neuropeptide Y receptor Y5 (NPY5R, NY5R)
- Prolactin-releasing peptide receptor (PRLHR, GPRA) InterPro: IPR001402
- Prokineticin receptor 1 (PROKR1, GPR73)
- Prokineticin receptor 2 (PROKR2, PKR2)
- GPR19 (GPR19, GPRJ)
- GPR50 (GPR50, ML1X)
- GPR75 (GPR75)
- GPR83 (GPR83, GPR72)
Subfamily A10
[edit]- Glycoprotein hormone receptor InterPro: IPR002131
- Leucine-rich repeat-containing G protein-coupled receptor 4 (LGR4, GPR48)
- Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5, GPR49)
- Leucine-rich repeat-containing G protein-coupled receptor 6 (LGR6)
Subfamily A11
[edit]- GPR40-related receptor InterPro: IPR013312
- Free fatty acid receptor 1 (FFAR1, GPR40)
- Free fatty acid receptor 2 (FFAR2, GPR43)
- Free fatty acid receptor 3 (FFAR3, GPR41)
- GPR42 (GPR42, FFAR1L)
- P2 purinoceptor InterPro: IPR002286
- Hydroxycarboxylic acid receptor 1 (HCAR1, GPR81)
- Hydroxycarboxylic acid receptor 2, Niacin receptor 1 (HCAR2, GPR109A)
- Hydroxycarboxylic acid receptor 3, Niacin receptor 2 (HCAR3, GPR109B, HM74)
- GPR31 (GPR31, GPRV)
- GPR82 (GPR82)
- Oxoglutarate (alpha-ketoglutarate) receptor 1 (OXGR1, GPR80)
- Succinate receptor 1 (SUCNR1, GPR91)
Subfamily A12
[edit]- P2 purinoceptor InterPro: IPR002286
- Purinergic receptor P2Y12 (P2RY12)
- Purinergic receptor P2Y13 (P2RY13, GPR86) InterPro: IPR008109
- Purinergic receptor P2Y14 (P2RY14, UDP-glucose receptor, KI01) InterPro: IPR005466
- GPR34 (GPR34)
- GPR87 (GPR87)
- GPR171 (GPR171, H963)
- Platelet-activating factor receptor (PTAFR, PAFR) InterPro: IPR002282
Subfamily A13
[edit]- Cannabinoid receptor InterPro: IPR002230
- Cannabinoid receptor 1 (brain) (CNR1, CB1R)
- Cannabinoid receptor 2 (macrophage) (CNR2, CB2R)
- Lysophosphatidic acid receptor InterPro: IPR004065
- Sphingosine 1-phosphate receptor InterPro: IPR004061
- Melanocortin/ACTH receptor InterPro: IPR001671
- Melanocortin 1 receptor (MC1R, MSHR)
- Melanocortin 3 receptor (MC3R)
- Melanocortin 4 receptor (MC4R)
- Melanocortin 5 receptor (MC5R)
- ACTH receptor (MC2R), ACTR)
- GPR3 (GPR3)
- GPR6 (GPR6)
- GPR12 (GPR12, GPRC)
Subfamily A14
[edit]- Eicosanoid receptor InterPro: IPR008365
- Prostaglandin D2 receptor (PTGDR, PD2R)
- Prostaglandin E1 receptor (PTGER1, PE21)
- Prostaglandin E2 receptor (PTGER2, PE22)
- Prostaglandin E3 receptor (PTGER3, PE23)
- Prostaglandin E4 receptor (PTGER4, PE24)
- Prostaglandin F receptor (PTGFR, PF2R)
- Prostaglandin I2 (prostacyclin) receptor (PTGIR, PI2R)
- Thromboxane A2 receptor (TBXA2R, TA2R)
Subfamily A15
[edit]- Lysophosphatidic acid receptor InterPro: IPR004065
- P2 purinoceptor InterPro: IPR002286
- Purinergic receptor P2Y10 (P2RY10, P2Y10)
- Protease-activated receptor InterPro: IPR003912
- Epstein-Barr virus induced gene 2 (lymphocyte-specific G protein-coupled receptor) (GPR183)
- Proton-sensing G protein-coupled receptors
- GPR17 (GPR17, GPRH)
- GPR18 (GPR18, GPRI)
- GPR20 (GPR20, GPRK)
- GPR35 (GPR35)
- GPR55 (GPR55)
- Coagulation factor II receptor (F2R, THRR)
Subfamily A16
[edit]- Opsins InterPro: IPR001760[2]
- Rhodopsin (RHO, OPSD)
- Opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) (OPN1SW, OPSB) (blue-sensitive opsin)
- Opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) (OPN1MW, OPSG) (green-sensitive opsin)
- Opsin 1 (cone pigments), long-wave-sensitive (color blindness, protan) (OPN1LW, OPSR) (red-sensitive opsin)
- Opsin 3, Panopsin (OPN3)
- Opsin 4, Melanopsin (OPN4)
- Opsin 5 (OPN5, GPR136)
- Retinal G protein coupled receptor (RGR)
- Retinal pigment epithelium-derived rhodopsin homolog (RRH, OPSX) (visual pigment-like receptor opsin) InterPro: IPR001793
Subfamily A17
[edit]- 5-Hydroxytryptamine (5-HT) receptor InterPro: IPR002231
- Adrenergic receptor InterPro: IPR002233
- Dopamine receptor InterPro: IPR000929
- Trace amine receptor InterPro: IPR009132
- Histamine H2 receptor (HRH2, HH2R) InterPro: IPR000503
Subfamily A18
[edit]- Histamine H1 receptor (HRH1, HH1R) InterPro: IPR000921
- Histamine H3 receptor (HRH3) InterPro: IPR003980
- Histamine H4 receptor (HRH4) InterPro: IPR008102
- Adenosine receptor InterPro: IPR001634
- Muscarinic acetylcholine receptor InterPro: IPR000995
- GPR21 (GPR21, GPRL)
- GPR27 (GPR27)
- GPR45 (GPR45, PSP24)
- GPR52 (GPR52)
- GPR61 (GPR61)
- GPR62 (GPR62)
- GPR63 (GPR63)
- GPR78 (GPR78)
- GPR84 (GPR84)
- GPR85 (GPR85)
- GPR88 (GPR88)
- GPR101 (GPR101)
- GPR161 (GPR161, RE2)
- GPR173 (GPR173, SREB3)
Subfamily A19
[edit]Unclassified
[edit]- Olfactory receptor InterPro: IPR000725
- Vomeronasal receptor InterPro: IPR004072
Secretin family of 7 transmembrane receptors This family is known as Family B, the secretin-receptor family or family 2 of the G-protein-coupled receptors (GPCR). Many secretin receptors are regulated by peptide hormones from the glucagon hormone family.
The secretin-receptor family GPCRs include
- vasoactive intestinal peptide receptors
- secretin,
- calcitonin and
- parathyroid hormone/parathyroid hormone-related peptides.
These receptors activate adenylyl cyclase and the phosphatidyl-inositol-calcium pathway.
The receptors in this family have 7 transmembrane helices,[3] like rhodopsin-like GPCRs. However, there is no significant sequence identity between these two GPCR families and the secretin-receptor family has its own characteristic 7TM signature.[4]
The secretin-receptor family GPCRs exist in many animal species and have not been identified in plants, fungi or prokaryotes. Three distinct sub-families (B1-B3) are recognized.
Subfamily B1
[edit]Subfamily B1 contains classical hormone receptors, such as receptors for secretin and glucagon, that are all involved in cAMP-mediated signalling pathways.
- Pituitary adenylate cyclase-activating polypeptide type 1 receptor InterPro: IPR002285
- Calcitonin receptor InterPro: IPR003287
- Corticotropin-releasing hormone receptor InterPro: IPR003051
- Glucose-dependent insulinotropic polypeptide receptor/Gastric inhibitory polypeptide receptor InterPro: IPR001749
- Glucagon receptor InterPro: IPR003291
- Glucagon receptor-related InterPro: IPR003290
- Growth hormone releasing hormone receptor InterPro: IPR003288
- Parathyroid hormone receptor InterPro: IPR002170
- Secretin receptor InterPro: IPR002144
- Vasoactive intestinal peptide receptor InterPro: IPR001571
Subfamily B2
[edit]Subfamily B2 contains receptors with long extracellular N-termini, such as the leukocyte cell-surface antigen CD97; calcium-independent receptors for latrotoxin (such as O94910, and brain-specific angiogenesis inhibitor receptors (such as O14514) amongst others.
- Brain-specific angiogenesis inhibitor InterPro: IPR008077
- CD97 antigen InterPro: IPR003056
- EMR hormone receptor InterPro: IPR001740
- GPR56 orphan receptor InterPro: IPR003910
- Latrophilin receptor InterPro: IPR003924
Subfamily B3
[edit]Subfamily B3 includes Methuselah and other Drosophila proteins. Other than the typical seven-transmembrane region, characteristic structural features include an amino-terminal extracellular domain involved in ligand binding, and an intracellular loop (IC3) required for specific G-protein coupling.
Unclassified subfamilies
[edit]Unclassified members
[edit]HCTR-5; HCTR-6; KPG 006; KPG 008
Metabotropic Glutamate/Pheromone receptors
[edit]Eight different types of mGluRs, labeled mGluR1 to mGluR8 (GRM1 to GRM8), are divided into groups I, II, and III.[5][6][7][8] Receptor types are grouped based on receptor structure and physiological activity.[9] The mGluRs are further divided into subtypes, such as mGluR7a and mGluR7b.
Overview
[edit]| Family | Receptors [10][11] | Gene | Mechanism[10] | Function | Agonists & Activators | Antagonists | Synapse site |
|---|---|---|---|---|---|---|---|
| Group I | mGluR1 | GRM1 | Gq, ↑Na+,[7] ↑K+,[7] ↓glutamate[8] |
|
|
mainly postsynaptic[14] | |
| mGluR5 | GRM5 | Gq, ↑Na+,[7] ↑K+,[7] ↓glutamate[8] | |||||
| Group II | mGluR2 | GRM2 | Gi/G0 |
|
|
|
mainly presynaptic[14] |
| mGluR3 | GRM3 | Gi/G0 | |||||
| Group III | mGluR4 | GRM4 | Gi/G0 |
|
|
mainly presynaptic[14] | |
| mGluR6 | GRM6 | Gi/G0 | |||||
| mGluR7 | GRM7 | Gi/G0 | |||||
| mGluR8 | GRM8 | Gi/G0 |
This receptor resembles many 7 transmembrane G-protiens found in humans, though it is know known at this time if human genetics code for this type of receptor
| Fungal pheromone mating factor STE2 GPCR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 Structure of a Peptide Segment of the 6th Transmembrane Domain of the Saccharomyces cerevisiae alpha-Factor Receptor.[16] | |||||||||||
| Identifiers | |||||||||||
| Symbol | STE2 | ||||||||||
| Pfam | PF02116 | ||||||||||
| InterPro | IPR000366 | ||||||||||
| SCOP2 | 1pjd / SCOPe / SUPFAM | ||||||||||
| OPM superfamily | 6 | ||||||||||
| OPM protein | 2k9p | ||||||||||
| |||||||||||
Fungal pheromone mating factor receptors form a distinct family of G-protein-coupled receptors.
Mating factor receptors STE2 and STE3 are integral membrane proteins that may be involved in the response to mating factors on the cell membrane.[17][18][19] The amino acid sequences of both receptors contain high proportions of hydrophobic residues grouped into 7 domains, in a manner reminiscent of the rhodopsins and other receptors believed to interact with G-proteins.
| Slime mold cyclic AMP receptor | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Dicty_CAR | ||||||||
| Pfam | PF05462 | ||||||||
| Pfam clan | CL0192 | ||||||||
| InterPro | IPR000848 | ||||||||
| PROSITE | PDOC00691 | ||||||||
| |||||||||
Cyclic AMP receptors from slime molds are a distinct family of G-protein coupled receptors. These receptors control development in Dictyostelium discoideum.
The cyclic AMP receptors coordinate aggregation of individual cells into a multicellular organism, and regulate the expression of a large number of developmentally-regulated genes.[20][21][22] The amino acid sequences of the receptors contain high proportions of hydrophobic residues grouped into 7 domains, in a manner reminiscent of the rhodopsins and other receptors believed to interact with G-proteins. However, while a similar 3D framework has been proposed to account for this, there is no significant sequence similarity between these families: the cAMP receptors thus bear their own unique '7TM' signature.
See also
[edit]Smoothened is a G protein-coupled receptor[23] protein encoded by the SMO gene of the hedgehog signaling pathway conserved from flies to humans. It is the molecular target of the teratogen cyclopamine.[24]
Cellular localization plays an essential role in the function of SMO. Stimulation of the patched receptor by the sonic hedgehog ligand leads to translocation of SMO to the primary cilium. Furthermore, SMO that is mutated in the domain required for ciliary localisation cannot contribute to pathway activation.[25] SMO has also been shown to bind the kinesin motor protein Costal-2 and play a role in the localization of the Ci (Cubitus interruptus transcription factor) complex.[26]
SMO can function as an oncogene. Activating SMO mutations can lead to unregulated activation of the hedgehog pathway and cancer.[27]
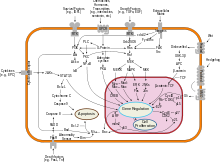
Agonists
[edit]Antagonists
[edit]- Cyclopamine
- Itraconazole
- Vismodegib (Erivedge), a smoothened receptor inhibitor for the treatment of basal-cell carcinoma, being investigated for the treatment of other types of cancer
Frizzled
[edit]| Frizzled/Smoothened family membrane region | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal structure of the cysteine-rich domain of mouse frizzled 8 (mfz8)[28] | |||||||||
| Identifiers | |||||||||
| Symbol | Frizzled | ||||||||
| Pfam | PF01534 | ||||||||
| Pfam clan | GPCR_A | ||||||||
| InterPro | IPR000539 | ||||||||
| PROSITE | PDOC50038 | ||||||||
| TCDB | 9.A.14 | ||||||||
| OPM superfamily | 6 | ||||||||
| OPM protein | 4jkv | ||||||||
| |||||||||
Frizzled is a family of G protein-coupled receptor proteins[29] that serves as receptors in the Wnt signaling pathway and other signaling pathways. When activated, Frizzled leads to activation of Dishevelled in the cytosol.
Species distribution
[edit]Frizzled proteins and the genes that encode them have been identified in an array of animals, from sponges to humans.[30]
Function
[edit]Frizzled proteins also play key roles in governing cell polarity, embryonic development, formation of neural synapses, cell proliferation, and many other processes in developing and adult organisms. These processes occur as a result of one of three signaling pathways. These include the canonical Wnt/β-catenin pathway, Wnt/calcium pathway, and planar cell polarity (PCP) pathway.[30] Mutations in the human frizzled-4 receptor have been linked to familial exudative vitreoretinopathy, a rare disease affecting the retina at the back of the eye, and the vitreous, the clear fluid inside the eye.
The frizzled (fz) locus of Drosophila coordinates the cytoskeletons of epidermal cells, producing a parallel array of cuticular hairs and bristles.[31][32] In fz mutants, the orientation of individual hairs with respect both to their neighbours and to the organism as a whole is altered. In the wild-type wing, all hairs point towards the distal tip.[32]
In the developing wing, Fz has 2 functions: it is required for the proximal-distal transmission of an intracellular polarity signal; and it is required for cells to respond to the polarity signal. Fz produces an mRNA that encodes an integral membrane protein with 7 putative transmembrane (TM) domains. This protein should contain both extracellular and cytoplasmic domains, which could function in the transmission and interpretation of polarity information.[32] This signature is usually found downstream of the Fz domain (InterPro: IPR000024)
=Cysteine-rich domain
[edit]Frizzled proteins include cysteine-rich domain that is conserved in diverse proteins, including several receptor tyrosine kinases.[33][34][35] In Drosophila melanogaster, members of the Frizzled family of tissue-polarity genes encode proteins that appear to function as cell-surface receptors for Wnts. The Frizzled genes belong to the seven transmembrane class of receptors (7TMR) and have in their extracellular region a cysteine-rich domain that has been implicated as the Wnt binding domain. Sequence similarity between the cysteine-rich domain of Frizzled and several receptor tyrosine kinases, which have roles in development, include the muscle-specific receptor tyrosine kinase (MuSK), the neuronal-specific kinase (NSK2), and ROR1 and ROR2. The structure of this domain is known and is composed mainly of alpha helices. This domain contains ten conserved cysteines that form five disulphide bridges.
Group members
[edit]The following is a list of the ten known human frizzled receptors:
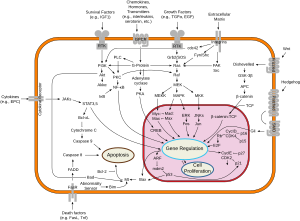
- Frizzled-1 (FZD1)
- Frizzled-2 (FZD2)
- Frizzled-3 (FZD3)
- Frizzled-4 (FZD4)
- Frizzled-5 (FZD5)
- Frizzled-6 (FZD6)
- Frizzled-7 (FZD7)
- Frizzled-8 (FZD8)
- Frizzled-9 (FZD9)
- Frizzled-10 (FZD10)
| receptor protein-tyrosine kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 2.7.10.1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Identifiers | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | ? | ||||||||
| Pfam | PF07714 | ||||||||
| OPM superfamily | 207 | ||||||||
| OPM protein | 2k1k | ||||||||
| |||||||||
Receptor tyrosine kinases (RTK)s are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. Of the 90 unique tyrosine kinase genes identified in the human genome, 58 encode receptor tyrosine kinase proteins.[36] Receptor tyrosine kinases have been shown not only to be key regulators of normal cellular processes but also to have a critical role in the development and progression of many types of cancer.[37] Receptor tyrosine kinases are part of the larger family of protein tyrosine kinases, encompassing the receptor tyrosine kinase proteins which contain a transmembrane domain, as well as the nonreceptor tyrosine kinases which do not possess transmembrane domains.[38]
Receptor tyrosine kinase (RTK) in this article is also known as tyrosine receptor kinase (TRK) or tyrosine kinase receptor (TKR) depending on the permutation. However, Trk receptor is the name in the context of neurobiology.
Receptor tyrosine kinase classes
[edit]Approximately 20 different RTK classes have been identified.[39]
- RTK class I (EGF receptor family) (ErbB family)
- RTK class II (Insulin receptor family)
- RTK class III (PDGF receptor family)
- RTK class IV (FGF receptor family)
- RTK class V (VEGF receptors family)
- RTK class VI (HGF receptor family)
- RTK class VII (Trk receptor family)
- RTK class VIII (Eph receptor family)
- RTK class IX (AXL receptor family)
- RTK class X (LTK receptor family)
- RTK class XI (TIE receptor family)
- RTK class XII (ROR receptor family)
- RTK class XIII (DDR receptor family)
- RTK class XIV (RET receptor family)
- RTK class XV (KLG receptor family)
- RTK class XVI (RYK receptor family)
- RTK class XVII (MuSK receptor family)
The main receptors in the immune system are pattern recognition receptors (PRRs), Toll-like receptors (TLRs), killer activated and killer inhibitor receptors (KARs and KIRs), complement receptors, Fc receptors, B cell receptors and T cell receptors.[40]
| Receptor | Bind to [40] | Function[40] |
|---|---|---|
| pattern recognition receptors (PRRs) (e.g. TLRs, NLRs) |
pathogen-associated molecular patterns (PAMP) | Mediate cytokine production --> inflammation --> destroying pathogen |
| killer activated and killer inhibitor receptors (KARs and KIRs) | Avails NK cells to identify abnormal host cells (KAR) or inhibit inappropriate host cell destruction (KIR) | |
| complement receptors | complement proteins on e.g. microbes | Allow phagocytic and B cells to recognize microbes and immune complexes |
| Fc receptors | epitope-antibody complexes | Stimulate phagocytosis |
| B cell receptors | epitopes | B cell differentiation into plasma cells and proliferation |
| T cell receptors | linear epitopes bound to MHC | Activate T cells |
| Cytokine receptors | cytokines | regulation and co-ordination of immune responses |
[[Nuclear receptors]]
[edit]The following is a list of the 48 known human nuclear receptors[41] categorized according to sequence homology.[42][43]
| Subfamily | Group | Member | ||||||
|---|---|---|---|---|---|---|---|---|
| NRNC Symbol[42] | Abbreviation | Name | Gene | Ligand(s) | ||||
| 1 | Thyroid Hormone Receptor-like | A | Thyroid hormone receptor | NR1A1 | TRα | Thyroid hormone receptor-α | THRA | thyroid hormone |
| NR1A2 | TRβ | Thyroid hormone receptor-β | THRB | |||||
| B | Retinoic acid receptor | NR1B1 | RARα | Retinoic acid receptor-α | RARA | vitamin A and related compounds | ||
| NR1B2 | RARβ | Retinoic acid receptor-β | RARB | |||||
| NR1B3 | RARγ | Retinoic acid receptor-γ | RARG | |||||
| C | Peroxisome proliferator-activated receptor | NR1C1 | PPARα | Peroxisome proliferator-activated receptor-α | PPARA | fatty acids, prostaglandins | ||
| NR1C2 | PPAR-β/δ | Peroxisome proliferator-activated receptor-β/δ | PPARD | |||||
| NR1C3 | PPARγ | Peroxisome proliferator-activated receptor-γ | PPARG | |||||
| D | Rev-ErbA | NR1D1 | Rev-ErbAα | Rev-ErbAα | NR1D1 | heme | ||
| NR1D2 | Rev-ErbAβ | Rev-ErbAα | NR1D2 | |||||
| F | RAR-related orphan receptor | NR1F1 | RORα | RAR-related orphan receptor-α | RORA | cholesterol, ATRA | ||
| NR1F2 | RORβ | RAR-related orphan receptor-β | RORB | |||||
| NR1F3 | RORγ | RAR-related orphan receptor-γ | RORC | |||||
| H | Liver X receptor-like | NR1H3 | LXRα | Liver X receptor-α | NR1H3 | oxysterols | ||
| NR1H2 | LXRβ | Liver X receptor-β | NR1H2 | |||||
| NR1H4 | FXR | Farnesoid X receptor | NR1H4 | |||||
| I | Vitamin D receptor-like | NR1I1 | VDR | Vitamin D receptor | VDR | vitamin D | ||
| NR1I2 | PXR | Pregnane X receptor | NR1I2 | xenobiotics | ||||
| NR1I3 | CAR | Constitutive androstane receptor | NR1I3 | androstane | ||||
| X | NRs with two DNA binding domains[44][45] | NR1X1 | 2DBD-NRα | |||||
| NR1X2 | 2DBD-NRβ | |||||||
| NR1X3 | 2DBD-NRγ | |||||||
| 2 | Retinoid X Receptor-like | A | Hepatocyte nuclear factor-4 | NR2A1 | HNF4α | Hepatocyte nuclear factor-4-α | HNF4A | fatty acids |
| NR2A2 | HNF4γ | Hepatocyte nuclear factor-4-γ | HNF4G | |||||
| B | Retinoid X receptor | NR2B1 | RXRα | Retinoid X receptor-α | RXRA | retinoids | ||
| NR2B2 | RXRβ | Retinoid X receptor-β | RXRB | |||||
| NR2B3 | RXRγ | Retinoid X receptor-γ | RXRG | |||||
| C | Testicular receptor | NR2C1 | TR2 | Testicular receptor 2 | NR2C1 | |||
| NR2C2 | TR4 | Testicular receptor 4 | NR2C2 | |||||
| E | TLX/PNR | NR2E1 | TLX | Homologue of the Drosophila tailless gene | NR2E1 | |||
| NR2E3 | PNR | Photoreceptor cell-specific nuclear receptor | NR2E3 | |||||
| F | COUP/EAR | NR2F1 | COUP-TFI | Chicken ovalbumin upstream promoter-transcription factor I | NR2F1 | |||
| NR2F2 | COUP-TFII | Chicken ovalbumin upstream promoter-transcription factor II | NR2F2 | |||||
| NR2F6 | EAR-2 | V-erbA-related | NR2F6 | |||||
| 3 | Estrogen Receptor-like | A | Estrogen receptor | NR3A1 | ERα | Estrogen receptor-α | ESR1 | estrogens |
| NR3A2 | ERβ | Estrogen receptor-β | ESR2 | |||||
| B | Estrogen related receptor | NR3B1 | ERRα | Estrogen-related receptor-α | ESRRA | |||
| NR3B2 | ERRβ | Estrogen-related receptor-β | ESRRB | |||||
| NR3B3 | ERRγ | Estrogen-related receptor-γ | ESRRG | |||||
| C | 3-Ketosteroid receptors | NR3C1 | GR | Glucocorticoid receptor | NR3C1 | cortisol | ||
| NR3C2 | MR | Mineralocorticoid receptor | NR3C2 | aldosterone | ||||
| NR3C3 | PR | Progesterone receptor | PGR | progesterone | ||||
| NR3C4 | AR | Androgen receptor | AR | testosterone | ||||
| 4 | Nerve Growth Factor IB-like | A | NGFIB/NURR1/NOR1 | NR4A1 | NGFIB | Nerve Growth factor IB | NR4A1 | |
| NR4A2 | NURR1 | Nuclear receptor related 1 | NR4A2 | |||||
| NR4A3 | NOR1 | Neuron-derived orphan receptor 1 | NR4A3 | |||||
| 5 | Steroidogenic Factor-like | A | SF1/LRH1 | NR5A1 | SF1 | Steroidogenic factor 1 | NR5A1 | phosphatidylinositols |
| NR5A2 | LRH-1 | Liver receptor homolog-1 | NR5A2 | phosphatidylinositols | ||||
| 6 | Germ Cell Nuclear Factor-like | A | GCNF | NR6A1 | GCNF | Germ cell nuclear factor | NR6A1 | |
| 0 | Miscellaneous | B | DAX/SHP | NR0B1 | DAX1 | Dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 | NR0B1 | |
| NR0B2 | SHP | Small heterodimer partner | NR0B2 | |||||
Typical Classifications of receptor families
[edit](gleanings from Receptor_(biochemistry)#Structure)
The structures of receptors are very diverse and can broadly be classified into the following categories:
These associated receptors are typical targets of fast neurotransmission or used for rapid signal transduction
Vertebrate Anionic Cys-loop Receptors
| Type | Class | IUPHAR-recommended protein name[46] |
Gene | Previous names |
|---|---|---|---|---|
| GABAA | alpha | α1 α2 α3 α4 α5 α6 |
GABRA1 GABRA2 GABRA3 GABRA4 GABRA5 GABRA6 |
EJM, ECA4 |
| beta | β1 β2 β3 |
GABRB1 GABRB2 GABRB3 |
ECA5 | |
| gamma | γ1 γ2 γ3 |
GABRG1 GABRG2 GABRG3 |
CAE2, ECA2, GEFSP3 | |
| delta | δ | GABRD | ||
| epsilon | ε | GABRE | ||
| pi | π | GABRP | ||
| theta | θ | GABRQ | ||
| rho | ρ1 ρ2 ρ3 |
GABRR1 GABRR2 GABRR3 |
GABAC[47] | |
| Glycine (GlyR) |
alpha | α1 α2 α3 α4 |
GLRA1 GLRA2 GLRA3 GLRA4 |
STHE |
| beta | β | GLRB |
Vertebrate Cationic Cys-loop Receptors
| Type | Class | IUPHAR-recommended protein name [46] |
Gene | Previous names |
|---|---|---|---|---|
| Serotonin (5-HT) |
5-HT3 | 5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
HTR3A HTR3B HTR3C HTR3D HTR3E |
5-HT3A 5-HT3B 5-HT3C 5-HT3D 5-HT3E |
| Nicotinic acetylcholine (nAChR) |
alpha | α1 α2 α3 α4 α5 α6 α7 α9 α10 |
CHRNA1 CHRNA2 CHRNA3 CHRNA4 CHRNA5 CHRNA6 CHRNA7 CHRNA9 CHRNA10 |
ACHRA, ACHRD, CHRNA, CMS2A, FCCMS, SCCMS |
| beta | β1 β2 β3 β4 |
CHRNB1 CHRNB2 CHRNB3 CHRNB4 |
CMS2A, SCCMS, ACHRB, CHRNB, CMS1D EFNL3, nAChRB2 | |
| gamma | γ | CHRNG | ACHRG | |
| delta | δ | CHRND | ACHRD, CMS2A, FCCMS, SCCMS | |
| epsilon | ε | CHRNE | ACHRE, CMS1D, CMS1E, CMS2A, FCCMS, SCCMS | |
| Zinc-activated ion channel (ZAC) |
ZAC | ZACN | ZAC1, L2m LGICZ, LGICZ1 |
Other ionotropic receptors:
- AMPA receptor
- Glycine receptor
- GRIN1
- GRIN3A
- GRIN3B
- GRINL1A
- GRINL1B
- HTR3A
- HTR3B
- HTR3C
- HTR3D
- HTR3E
- Kainate receptor
- NMDA receptor
- P2X purinoreceptor
in other (nonhuman) organisms
Activation of these receptor results in changes in ion movement across the membrane. They have a hetero structure. Each subunit consists of the extracellular ligand-binding domain and a transmembrane domain where the transmembrane domain in turn includes four transmembrane alpha helixes. The ligand binding cavities are located at the interface between the subunits.
G protein-coupled receptors (metabotropic)
[edit]GPCRs are involved in a wide variety of physiological processes. Some examples of their physiological roles include:
- The visual sense: The opsins use a photoisomerization reaction to translate electromagnetic radiation into cellular signals. Rhodopsin, for example, uses the conversion of 11-cis-retinal to all-trans-retinal for this purpose
- The gustatory sense (taste): GPCRs in taste cells mediate release of gustducin in response to bitter- and sweet-tasting substances.
- The sense of smell: Receptors of the olfactory epithelium bind odorants (olfactory receptors) and pheromones (vomeronasal receptors)
- Behavioral and mood regulation: Receptors in the mammalian brain bind several different neurotransmitters, including serotonin, dopamine, GABA, and glutamate
- Regulation of immune system activity and inflammation: Chemokine receptors bind ligands that mediate intercellular communication between cells of the immune system; receptors such as histamine receptors bind inflammatory mediators and engage target cell types in the inflammatory response
- Autonomic nervous system transmission: Both the sympathetic and parasympathetic nervous systems are regulated by GPCR pathways, responsible for control of many automatic functions of the body such as blood pressure, heart rate, and digestive processes
- Cell density sensing: A novel GPCR role in regulating cell density sensing.
- Homeostasis modulation (e.g., water balance).[48]
- Involved in growth and metastasis of some types of tumors.[49]
They are the largest family of receptors including the receptors for several hormones and slow transmitters e.g. dopamine, metabotropic glutamate, olfactory sense, etc.
Composed of seven transmembrane alpha helices. The loops connecting the alpha helices form extracellular and intracellular domains. The binding site for larger peptidic ligands is usually located in the extracellular domain whereas the binding site for smaller non-peptidic ligands is often located between the seven alpha helices and one extracellular loop.[50] These receptors are coupled to different intracellular effector systems via G-proteins.
These receptors are composed of an extracellular domain containing the ligand binding site and an intracellular domain, often with enzymatic function, linked by a single transmembrane alpha helix. e.g. the insulin receptor.
Immune receptors function to recognize pathogens and induce or inhibit certain immune responses
While they are called Nuclear receptors, these are actually located in the cytosol and migrate to the nucleus after binding with their ligands. They are composed of a C-terminal ligand binding region, a core DNA-binding domain (DBD) and an N-terminal domain that contains the AF1(activation function 1) region. The core region has two zinc fingers that are responsible for recognising the DNA sequences specific to this receptor. The N-terminal interacts with other cellular transcription factors in a ligand independent manner and depending on these interactions it can modify the binding/activity of the receptor. Steroid and thyroid hormone receptors are examples of such receptors.[51]
- ^ Choe S (February 2002). "Potassium channel structures". Nat. Rev. Neurosci. 3 (2): 115–21. doi:10.1038/nrn727. PMID 11836519.
- ^ Terakita A (2005). "The opsins". Genome Biol. 6 (3): 213. doi:10.1186/gb-2005-6-3-213. PMC 1088937. PMID 15774036.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ PDB: 4L6R; Siu FY, He M, de Graaf C, Han GW, Yang D, Zhang Z, Zhou C, Xu Q, Wacker D, Joseph JS, Liu W, Lau J, Cherezov V, Katritch V, Wang M-W, Stevens RC (July 2013). "Structure of the human glucagon class B G-protein-coupled receptor". Nature. 499 (7459): 444–449. doi:10.1038/nature12393. PMID 23863937.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC (2014). "Insights into the structure of class B GPCRs". Trends Pharmacol Sci. 35 (1): 12–22. doi:10.1016/j.tips.2013.11.001. PMID 24359917.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cite error: The named reference
Bonsiwas invoked but never defined (see the help page). - ^ Cite error: The named reference
Hinoiwas invoked but never defined (see the help page). - ^ a b c d e Cite error: The named reference
Chu and Hablitzwas invoked but never defined (see the help page). - ^ a b c Cite error: The named reference
Endohwas invoked but never defined (see the help page). - ^ Cite error: The named reference
Ohashiwas invoked but never defined (see the help page). - ^ a b If not otherwise specified in table:TABLE 1 Classification of the metabotropic glutamate (mGlu) receptors From the following article:
- ^ Swanson CJ, Bures M, Johnson MP, Linden AM, Monn JA, Schoepp DD (2005). "Metabotropic glutamate receptors as novel targets for anxiety and stress disorders". Nature Reviews Drug Discovery. 4 (2): 131–44. doi:10.1038/nrd1630. PMID 15665858.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cite error: The named reference
pmid11378156was invoked but never defined (see the help page). - ^ Cite error: The named reference
pmid12117582was invoked but never defined (see the help page). - ^ a b c Cite error: The named reference
Shigemotowas invoked but never defined (see the help page). - ^ a b Cite error: The named reference
Ambrosiniwas invoked but never defined (see the help page). - ^ Valentine KG, Liu SF, Marassi FM; et al. (October 2001). "Structure and topology of a peptide segment of the 6th transmembrane domain of the Saccharomyces cerevisae alpha-factor receptor in phospholipid bilayers". Biopolymers. 59 (4): 243–56. doi:10.1002/1097-0282(20011005)59:4<243::AID-BIP1021>3.0.CO;2-H. PMID 11473349.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Nakayama N, Miyajima A, Arai K (1985). "Nucleotide sequences of STE2 and STE3, cell type-specific sterile genes from Saccharomyces cerevisiae". EMBO J. 4 (10): 2643–2648. PMC 554555. PMID 16453635.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Burkholder AC, Hartwell LH (1985). "The yeast alpha-factor receptor: structural properties deduced from the sequence of the STE2 gene". Nucleic Acids Res. 13 (23): 8463–8475. doi:10.1093/nar/13.23.8463. PMC 322145. PMID 3001640.
- ^ Herskowitz I, Marsh L (1988). "STE2 protein of Saccharomyces kluyveri is a member of the rhodopsin/beta-adrenergic receptor family and is responsible for recognition of the peptide ligand alpha factor". Proc. Natl. Acad. Sci. U.S.A. 85 (11): 3855–3859. doi:10.1073/pnas.85.11.3855. PMC 280318. PMID 2836861.
- ^ Devreotes PN, Kimmel AR, Johnson RL, Klein PS, Sun TJ, Saxe III CL (1988). "A chemoattractant receptor controls development in Dictyostelium discoideum". Science. 241 (4872): 1467–1472. doi:10.1126/science.3047871. PMID 3047871.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ginsburg GT, Louis JM, Johnson R, Devreotes PN, Kimmel AR, Saxe III CL (1993). "CAR2, a prestalk cAMP receptor required for normal tip formation and late development of Dictyostelium discoideum". Genes Dev. 7 (2): 262–272. doi:10.1101/gad.7.2.262. PMID 8436297.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Devreotes PN, Kimmel AR, Johnson RL, Gollop R, Saxe III CL (1993). "Identification and targeted gene disruption of cAR3, a cAMP receptor subtype expressed during multicellular stages of Dictyostelium development". Genes Dev. 7 (2): 273–282. doi:10.1101/gad.7.2.273. PMID 8382181.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ruiz-Gómez A, Molnar C, Holguín H, Mayor F, de Celis JF (2007). "The cell biology of Smo signalling and its relationships with GPCRs". Biochim. Biophys. Acta. 1768 (4): 901–12. doi:10.1016/j.bbamem.2006.09.020. PMID 17094938.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA (2000). "Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine". Nature. 406 (6799): 1005–9. doi:10.1038/35023008. PMID 10984056.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF (2005). "Vertebrate Smoothened functions at the primary cilium". Nature. 437 (7061): 1018–21. doi:10.1038/nature04117. PMID 16136078.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, Weis-Garcia F, Gong R, Wang B, Beachy PA (November 2003). "Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2". Mol. Cell. 12 (5): 1261–74. doi:10.1016/S1097-2765(03)00426-X. PMID 14636583.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, Rosenthal A, Epstein EH, de Sauvage FJ (1998). "Activating Smoothened mutations in sporadic basal-cell carcinoma". Nature. 391 (6662): 90–2. doi:10.1038/34201. PMID 9422511.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ PDB: 1IJY; Dann CE, Hsieh JC, Rattner A, Sharma D, Nathans J, Leahy DJ (July 2001). "Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains". Nature. 412 (6842): 86–90. doi:10.1038/35083601. PMID 11452312.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Malbon CC (2004). "Frizzleds: new members of the superfamily of G-protein-coupled receptors". Front. Biosci. 9: 1048–58. doi:10.2741/1308. PMID 14977528.
- ^ a b Huang HC, Klein PS (2004). "The Frizzled family: receptors for multiple signal transduction pathways". Genome Biol. 5 (7): 234. doi:10.1186/gb-2004-5-7-234. PMC 463283. PMID 15239825.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Adler PN, Vinson C, Park WJ, Conover S, Klein L (1990). "Molecular structure of frizzled, a Drosophila tissue polarity gene". Genetics. 126 (2): 401–16. PMC 1204194. PMID 2174014.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Adler PN, Conover S, Vinson CR (1989). "A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains". Nature. 338 (6212): 263–264. doi:10.1038/338263a0. PMID 2493583.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nusse R, Xu YK (1998). "The Frizzled CRD domain is conserved in diverse proteins including several receptor tyrosine kinases". Curr. Biol. 8 (12): R405–R406. doi:10.1016/S0960-9822(98)70262-3. PMID 9637908.
- ^ Saldanha J, Singh J, Mahadevan D (1998). "Identification of a Frizzled-like cysteine rich domain in the extracellular region of developmental receptor tyrosine kinases". Protein Sci. 7 (7): 1632–1635. doi:10.1002/pro.5560070718. PMC 2144063. PMID 9684897.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hofmann K, Pihlajaniemi T, Bucher P, Rehn M (1998). "The frizzled motif: in how many different protein families does it occur?". Trends Biochem. Sci. 23 (11): 415–417. doi:10.1016/S0968-0004(98)01290-0. PMID 9852758.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Robinson DR, Wu YM, Lin SF (November 2000). "The protein tyrosine kinase family of the human genome". Oncogene. 19 (49): 5548–57. doi:10.1038/sj.onc.1203957. PMID 11114734.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zwick E, Bange J, Ullrich A (September 2001). "Receptor tyrosine kinase signalling as a target for cancer intervention strategies". Endocr. Relat. Cancer. 8 (3): 161–73. doi:10.1677/erc.0.0080161. PMID 11566607.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hubbard SR, Till JH (2000). "Protein tyrosine kinase structure and function". Annu. Rev. Biochem. 69: 373–98. doi:10.1146/annurev.biochem.69.1.373. PMID 10966463.
- ^ http://www.genome.ad.jp [1] Retrieved on 2007-04-05
- ^ a b c Lippincott's Illustrated Reviews: Immunology. Paperback: 384 pages. Publisher: Lippincott Williams & Wilkins; (July 1, 2007). Language: English. ISBN 0-7817-9543-5. ISBN 978-0-7817-9543-2. Page 20
- ^ Cite error: The named reference
Zhang_2004was invoked but never defined (see the help page). - ^ a b Cite error: The named reference
NRNC_1999was invoked but never defined (see the help page). - ^ Cite error: The named reference
Laudet_1997was invoked but never defined (see the help page). - ^ Cite error: The named reference
Wu_2006was invoked but never defined (see the help page). - ^ Cite error: The named reference
Wu_2007was invoked but never defined (see the help page). - ^ a b Collingridge GL, Olsen RW, Peters J, Spedding M (January 2009). "A nomenclature for ligand-gated ion channels". Neuropharmacology. 56 (1): 2–5. doi:10.1016/j.neuropharm.2008.06.063. PMC 2847504. PMID 18655795.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Olsen RW, Sieghart W (September 2008). "International Union of Pharmacology. LXX. Subtypes of γ-Aminobutyric AcidA Receptors: Classification on the Basis of Subunit Composition, Pharmacology, and Function. Update". Pharmacol. Rev. 60 (3): 243–60. doi:10.1124/pr.108.00505. PMC 2847512. PMID 18790874.
- ^ Hazell GG, Hindmarch CC, Pope GR, Roper JA, Lightman SL, Murphy D, O'Carroll AM, Lolait SJ (July 2011). "G protein-coupled receptors in the hypothalamic paraventricular and supraoptic nuclei - serpentine gateways to neuroendocrine homeostasis". Front Neuroendocrinol. 33 (1): 45–66. doi:10.1016/j.yfrne.2011.07.002. PMC 3336209. PMID 21802439.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Dorsam RT, Gutkind JS. (Feb 2007). "G-protein-coupled receptors and cancer". Nat Rev Cancer. 7 (2): 79–94. doi:10.1038/nrc2069. PMID 17251915.
- ^ Congreve M, Marshall F (March 2010). "The impact of GPCR structures on pharmacology and structure-based drug design". Br. J. Pharmacol. 159 (5): 986–96. doi:10.1111/j.1476-5381.2009.00476.x. PMC 2839258. PMID 19912230.
- ^ Rang HP, Dale MM, Ritter JM, Flower RJ, Henderson G (2012). Rang & Dale's Pharmacology (7th ed.). Elsevier Churchill Livingstone. ISBN 978-0-7020-3471-8.
{{cite book}}: CS1 maint: multiple names: authors list (link)
