Protein kinase domain
| Protein kinase domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
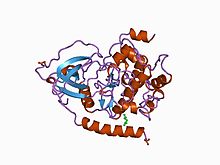 Structure of the catalytic subunit of cAMP-dependent protein kinase.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Pkinase | ||||||||
| Pfam | PF00069 | ||||||||
| InterPro | IPR000719 | ||||||||
| SMART | TyrKc | ||||||||
| PROSITE | PDOC00100 | ||||||||
| SCOP2 | 1apm / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 186 | ||||||||
| CDD | cd00180 | ||||||||
| Membranome | 3 | ||||||||
| |||||||||
The protein kinase domain is a structurally conserved protein domain containing the catalytic function of protein kinases.[2][3][4] Protein kinases are a group of enzymes that move a phosphate group onto proteins, in a process called phosphorylation. This functions as an on/off switch for many cellular processes, including metabolism, transcription, cell cycle progression, cytoskeletal rearrangement and cell movement, apoptosis, and differentiation. They also function in embryonic development, physiological responses, and in the nervous and immune system. Abnormal phosphorylation causes many human diseases, including cancer, and drugs that affect phosphorylation can treat those diseases.[5]
Protein kinases possess a catalytic subunit which transfers the gamma phosphate from nucleoside triphosphates (almost always ATP) to the side chain of an amino acid in a protein, resulting in a conformational and/or dynamic changes affecting protein function. These enzymes fall into two broad classes, characterised with respect to substrate specificity: serine/threonine specific and tyrosine specific.[6]
Function
[edit]Protein kinase function has been evolutionarily conserved from Escherichia coli to Homo sapiens. Protein kinases play a role in a multitude of cellular processes, including division, proliferation, apoptosis, and differentiation.[7] Phosphorylation usually results in a functional change of the target protein by changing structure, dynamics, enzyme activity, cellular location, or association with other proteins.
Structure
[edit]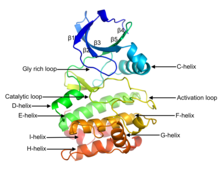
The catalytic subunits of protein kinases are highly conserved, and the structures of over 280 of the approximately 500 human kinase domains have been determined,[8] leading to large screens to develop kinase-specific inhibitors for the treatments of a number of diseases.[9] Humans have only 437 kinase domains that have catalytic activity; the rest are pseudokinases or catalyze other reactions.[citation needed]
Eukaryotic protein kinases[2][3][10][11] are enzymes that belong to a very extensive family of proteins which share a conserved catalytic core common with both serine/threonine and tyrosine protein kinases. The domain consists of two sub-domains referred to as the N- and C-terminal domains. The N-terminal domain consists of five beta sheet strands and an alpha helix called the C-helix, and the C-terminal domain usually consists of six alpha helices (labeled D, E, F, G, H, and I). The C-terminal domain contains two long loops, called the catalytic loop and the activation loop, which are essential for catalytic activity. The catalytic loop includes the "HRD motif" (for the amino acid sequence His-Arg-Asp), whose aspartic acid residue interacts directly with the hydroxyl group of the target serine, threonine, or tyrosine residue that is phosphorylated.[12]
The activation loop starts with the DFG motif (for the amino acid sequence Asp-Phe-Gly), which helps to bind ATP and magnesium in the active site. Broadly, the state or conformation of the kinase may be classified as DFGin or DFGout, depending on whether the Asp residue of the DFG motif is in or out of the active site. In the active form, the first few residues of the activation loop adopt a specific form of the DFGin conformation. Some inactive structures may adopt one of several other DFGin conformations, while other inactive structures are DFGout.[13]
Examples
[edit]The following is a list of human proteins containing the protein kinase domain:[14]
AAK1 ; AATK ; ABL1 ; ABL2 ; ACVR1 ; ACVR1B ; ACVR1C ; ACVR2A ; ACVR2B ; ACVRL1 ; AKT1 ; AKT2 ; AKT3 ; ALK ; AMHR2 ; ANKK1 ; ARAF ; AURKA ; AURKB ; AURKC ; AXL ; BLK ; BMP2K ; BMPR1A ; BMPR1B ; BMPR2 ; BMX ; BRAF ; BRSK1 ; BRSK2 ; BTK ; BUB1 ; BUB1B ; CAMK1 ; CAMK1D ; CAMK1G ; CAMK2A ; CAMK2B ; CAMK2D ; CAMK2G ; CAMK4 ; CAMKK1 ; CAMKK2 ; CAMKV ; CASK ; CDC42BPA ; CDC42BPB ; CDC42BPG ; CDC7 ; CDK1 ; CDK10 ; CDK11A ; CDK11B ; CDK12 ; CDK13 ; CDK14 ; CDK15 ; CDK16 ; CDK17 ; CDK18 ; CDK19 ; CDK2 ; CDK20 ; CDK3 ; CDK4 ; CDK5 ; CDK6 ; CDK7 ; CDK8 ; CDK9 ; CDKL1 ; CDKL2 ; CDKL3 ; CDKL4 ; CDKL5 ; CHEK1 ; CHEK2 ; CHUK ; CIT ; CLK1 ; CLK2 ; CLK3 ; CLK4 ; CSF1R ; CSK ; CSNK1A1 ; CSNK1A1L ; CSNK1D ; CSNK1E ; CSNK1G1 ; CSNK1G2 ; CSNK1G3 ; CSNK2A1 ; CSNK2A2 ; CSNK2A3 ; DAPK1 ; DAPK2 ; DAPK3 ; DCLK1 ; DCLK2 ; DCLK3 ; DDR1 ; DDR2 ; DMPK ; DSTYK ; DYRK1A ; DYRK1B ; DYRK2 ; DYRK3 ; DYRK4 ; EGFR ; EIF2AK1 ; EIF2AK2 ; EIF2AK3 ; EIF2AK4 ; EPHA1 ; EPHA10 ; EPHA2 ; EPHA3 ; EPHA4 ; EPHA5 ; EPHA6 ; EPHA7 ; EPHA8 ; EPHB1 ; EPHB2 ; EPHB3 ; EPHB4 ; EPHB6 ; ERBB2 ; ERBB3 ; ERBB4 ; ERN1 ; ERN2 ; FER ; FES ; FGFR1 ; FGFR2 ; FGFR3 ; FGFR4 ; FGR ; FLT1 ; FLT3 ; FLT4 ; FRK ; FYN ; GAK ; GRK1 ; GRK2 ; GRK3 ; GRK4 ; GRK5 ; GRK6 ; GRK7 ; GSG2 ; GSK3A ; GSK3B ; GUCY2C ; GUCY2D ; GUCY2F ; HCK ; HIPK1 ; HIPK2 ; HIPK3 ; HIPK4 ; HUNK ; ICK ; IGF1R ; IKBKB ; IKBKE ; ILK ; INSR ; INSRR ; IRAK1 ; IRAK2 ; IRAK3 ; IRAK4 ; ITK ; JAK1 ; JAK2 ; JAK3 ; KALRN ; KDR ; KIT ; KSR1 ; KSR2 ; LATS1 ; LATS2 ; LCK ; LIMK1 ; LIMK2 ; LMTK2 ; LMTK3 ; LRRK1 ; LRRK2 ; LTK ; LYN ; MAK ; MAP2K1 ; MAP2K2 ; MAP2K3 ; MAP2K4 ; MAP2K5 ; MAP2K6 ; MAP2K7 ; MAP3K1 ; MAP3K10 ; MAP3K11 ; MAP3K12 ; MAP3K13 ; MAP3K14 ; MAP3K15 ; MAP3K19 ; MAP3K2 ; MAP3K20 ; MAP3K21 ; MAP3K3 ; MAP3K4 ; MAP3K5 ; MAP3K6 ; MAP3K7 ; MAP3K8 ; MAP3K9 ; MAP4K1 ; MAP4K2 ; MAP4K3 ; MAP4K4 ; MAP4K5 ; MAPK1 ; MAPK10 ; MAPK11 ; MAPK12 ; MAPK13 ; MAPK14 ; MAPK15 ; MAPK3 ; MAPK4 ; MAPK6 ; MAPK7 ; MAPK8 ; MAPK9 ; MAPKAPK2 ; MAPKAPK3 ; MAPKAPK5 ; MARK1 ; MARK2 ; MARK3 ; MARK4 ; MAST1 ; MAST2 ; MAST3 ; MAST4 ; MASTL ; MATK ; MELK ; MERTK ; MET ; MINK1 ; MKNK1 ; MKNK2 ; MLKL ; MOK ; MOS ; MST1R ; MUSK ; MYLK ; MYLK2 ; MYLK3 ; MYLK4 ; MYO3A ; MYO3B ; NEK1 ; NEK10 ; NEK11 ; NEK2 ; NEK3 ; NEK4 ; NEK5 ; NEK6 ; NEK7 ; NEK8 ; NEK9 ; NIM1K ; NLK ; NPR1 ; NPR2 ; NRBP1 ; NRBP2 ; NRK ; NTRK1 ; NTRK2 ; NTRK3 ; NUAK1 ; NUAK2 ; OBSCN ; OXSR1 ; PAK1 ; PAK2 ; PAK3 ; PAK4 ; PAK5 ; PAK6 ; PAN3 ; PASK ; PBK ; PDGFRA ; PDGFRB ; PDIK1L ; PDPK1 ; PDPK2P ; PEAK1 ; PEAK3 ; PHKG1 ; PHKG2 ; PIK3R4 ; PIM1 ; PIM2 ; PIM3 ; PINK1 ; PKDCC ; PKMYT1 ; PKN1 ; PKN2 ; PKN3 ; PLK1 ; PLK2 ; PLK3 ; PLK4 ; PLK5 ; PNCK ; POMK ; PRKAA1 ; PRKAA2 ; PRKACA ; PRKACB ; PRKACG ; PRKCA ; PRKCB ; PRKCD ; PRKCE ; PRKCG ; PRKCH ; PRKCI ; PRKCQ ; PRKCZ ; PRKD1 ; PRKD2 ; PRKD3 ; PRKG1 ; PRKG2 ; PRKX ; PRKY ; PRPF4B ; PSKH1 ; PSKH2 ; PTK2 ; PTK2B ; PTK6 ; PTK7 ; PXK ; RAF1 ; RET ; RIOK1 ; RIOK2 ; RIOK3 ; RIPK1 ; RIPK2 ; RIPK3 ; RIPK4 ; RNASEL ; ROCK1 ; ROCK2 ; ROR1 ; ROR2 ; ROS1 ; RPS6KA1 ; RPS6KA2 ; RPS6KA3 ; RPS6KA4 ; RPS6KA5 ; RPS6KA6 ; RPS6KB1 ; RPS6KB2 ; RPS6KC1 ; RPS6KL1 ; RSKR ; RYK ; SBK1 ; SBK2 ; SBK3 ; SCYL1 ; SCYL2 ; SCYL3 ; SGK1 ; SGK2 ; SGK223 ; SGK3 ; SIK1 ; SIK1B ; SIK2 ; SIK3 ; SLK ; SNRK ; SPEG ; SRC ; SRMS ; SRPK1 ; SRPK2 ; SRPK3 ; STK10 ; STK11 ; STK16 ; STK17A ; STK17B ; STK24 ; STK25 ; STK26 ; STK3 ; STK31 ; STK32A ; STK32B ; STK32C ; STK33 ; STK35 ; STK36 ; STK38 ; STK38L ; STK39 ; STK4 ; STK40 ; STKLD1 ; STRADA ; STRADB ; STYK1 ; SYK ; TAOK1 ; TAOK2 ; TAOK3 ; TBCK ; TBK1 ; TEC ; TEK ; TESK1 ; TESK2 ; TEX14 ; TGFBR1 ; TGFBR2 ; TIE1 ; TLK1 ; TLK2 ; TNIK ; TNK1 ; TNK2 ; TNNI3K ; TP53RK ; TRIB1 ; TRIB2 ; TRIB3 ; TRIO ; TSSK1B ; TSSK2 ; TSSK3 ; TSSK4 ; TSSK6 ; TTBK1 ; TTBK2 ; TTK ; TTN ; TXK ; TYK2 ; TYRO3 ; UHMK1 ; ULK1 ; ULK2 ; ULK3 ; ULK4 ; VRK1 ; VRK2 ; VRK3 ; WEE1 ; WEE2 ; WNK1 ; WNK2 ; WNK3 ; WNK4 ; YES1 ; ZAP70
References
[edit]- ^ Knighton DR, Bell SM, Zheng J, et al. (May 1993). "2.0 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with a peptide inhibitor and detergent". Acta Crystallogr. D. 49 (Pt 3): 357–61. Bibcode:1993AcCrD..49..357K. doi:10.1107/S0907444993000502. PMID 15299526.
- ^ a b Hanks SK, Quinn AM (1991). "Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members". Protein Phosphorylation Part A: Protein Kinases: Assays, Purification, Antibodies, Functional Analysis, Cloning, and Expression. Methods in Enzymology. Vol. 200. pp. 38–62. doi:10.1016/0076-6879(91)00126-H. ISBN 978-0-12-182101-2. PMID 1956325.
- ^ a b Hanks SK, Hunter T (May 1995). "Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification". FASEB J. 9 (8): 576–96. doi:10.1096/fasebj.9.8.7768349. PMID 7768349. S2CID 21377422.
- ^ Scheeff ED, Bourne PE (October 2005). "Structural evolution of the protein kinase-like superfamily". PLOS Comput. Biol. 1 (5): e49. Bibcode:2005PLSCB...1...49S. doi:10.1371/journal.pcbi.0010049. PMC 1261164. PMID 16244704.
- ^ Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (December 2002). "The Protein Kinase Complement of the Human Genome". Science. 298 (5600): 1912–1934. Bibcode:2002Sci...298.1912M. doi:10.1126/science.1075762. PMID 12471243. S2CID 26554314.
- ^ Hunter T, Hanks SK, Quinn AM (1988). "The protein kinase family: conserved features and deduced phylogeny of the catalytic domains". Science. 241 (4861): 42–51. Bibcode:1988Sci...241...42H. doi:10.1126/science.3291115. PMID 3291115.
- ^ Manning G, Plowman GD, Hunter T, Sudarsanam S (October 2002). "Evolution of protein kinase signaling from yeast to man". Trends Biochem. Sci. 27 (10): 514–20. doi:10.1016/S0968-0004(02)02179-5. PMID 12368087.
- ^ Modi, V; Dunbrack, RL (24 December 2019). "A Structurally-Validated Multiple Sequence Alignment of 497 Human Protein Kinase Domains". Scientific Reports. 9 (1): 19790. Bibcode:2019NatSR...919790M. doi:10.1038/s41598-019-56499-4. PMC 6930252. PMID 31875044.
- ^ Li B, Liu Y, Uno T, Gray N (August 2004). "Creating chemical diversity to target protein kinases". Comb. Chem. High Throughput Screen. 7 (5): 453–72. doi:10.2174/1386207043328580. PMID 15320712. Archived from the original on 14 April 2013.
{{cite journal}}: CS1 maint: unfit URL (link) - ^ Hanks SK (2003). "Genomic analysis of the eukaryotic protein kinase superfamily: a perspective". Genome Biol. 4 (5): 111. doi:10.1186/gb-2003-4-5-111. PMC 156577. PMID 12734000.
- ^ Hunter T (1991). "Protein kinase classification". Protein Phosphorylation Part A: Protein Kinases: Assays, Purification, Antibodies, Functional Analysis, Cloning, and Expression. Methods in Enzymology. Vol. 200. pp. 3–37. doi:10.1016/0076-6879(91)00125-G. ISBN 978-0-12-182101-2. PMID 1835513.
- ^ Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM (July 1991). "Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase". Science. 253 (5018): 407–14. Bibcode:1991Sci...253..407K. doi:10.1126/science.1862342. PMID 1862342.
- ^ Modi, V; Dunbrack, RL (2 April 2019). "Defining a new nomenclature for the structures of active and inactive kinases". Proceedings of the National Academy of Sciences of the United States of America. 116 (14): 6818–6827. Bibcode:2019PNAS..116.6818M. doi:10.1073/pnas.1814279116. PMC 6452665. PMID 30867294.
- ^ "Human and mouse protein kinases: classification and index". pkinfam.txt. UniProt Consortium. Retrieved 10 June 2019.
