Talk:Variants of SARS-CoV-2/Archive 1
| This is an archive of past discussions about Variants of SARS-CoV-2. Do not edit the contents of this page. If you wish to start a new discussion or revive an old one, please do so on the current talk page. |
| Archive 1 |
Requested move 1 January 2021
The following discussion is closed. Please do not modify it. Subsequent comments should be made on the appropriate discussion page. No further edits should be made to this discussion.
Variants of severe acute respiratory syndrome coronavirus 2 → Variants of SARS-CoV-2 – Per [1]: SARS-CoV-2 (exact capitalisation and punctuation) is preferable [...] in the title of all other articles/category pages/etc. [than Severe acute respiratory syndrome coronavirus 2]
. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 00:35, 1 January 2021 (UTC)
- Hey, @Biscuit-in-Chief: Just wanted to let you know that I moved your move proposal on WP:Requested moves/Current discussions to this talk page. Per the instructions on WP:RM: When making a requested move proposal, you're supposed to create it on the talk page of the article to be moved, and a bot automatically adds the request to WP:Requested moves/Current discussions. Paintspot Infez (talk) 00:44, 1 January 2021 (UTC)
- Strong Support, this should be done ASAP to comply with consensus. I see no reason to have this article be any different. JackFromReedsburg (talk | contribs) 00:59, 1 January 2021 (UTC)
- Support. And sorry about that, quite new to article moving :-). —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 01:23, 1 January 2021 (UTC)
- Support. –Novem Linguae (talk) 03:05, 1 January 2021 (UTC)
- Support. —ACLNM (talk) 06:50, 1 January 2021 (UTC)
- Support. The spelt out Severe acute respiratory syndrome coronavirus 2 is appropriate for the parent article, but for derivative articles like this, the abbreviated form is better. —SmokeyJoe (talk) 10:26, 1 January 2021 (UTC)
- Support per argument above. 36.76.232.198 (talk) 23:58, 1 January 2021 (UTC)
Variant vs. strain
There is no universally accepted definition for the terms "strain", "variant", and "isolate" in the virology community, and most virologists simply copy the usage of terms from others. [...] According to Van Regenmortel, a (natural) virus strain is a "variant of a given virus that is recognizable because it possesses some unique phenotypic characteristics that remain stable under natural conditions" [emphasis added by the authors (van Regenmortel et al.)].[1] Such "unique phenotypic characteristics" are biological properties different from the compared reference virus, such as unique antigenic properties, host range or the signs of disease it causes. Importantly, as Van Regenmortel points out, a virus variant with a simple "difference in genome sequence…is not given the status of a separate strain since there is no recognizable distinct viral phenotype" [...].[2]
As of late December 2020, Talk:Variant of Concern 202012/01 § Please do not call it a strain. (variant, mutation, change) seems to suggest that general consensus on Wikipedia (although there is not much of it on this topic) is that "variant" is preferred for very specific subtypes of SARS-CoV-2—i.e., the ones in this category.[3]
References
- ^ van Regenmortel, M.H.V. (2007). "Virus species and virus identification: past and current controversies". Infection, Genetics and Evolution. 7 (1): 133–144. doi:10.1016/S1473-3099(11)70271-7. PMID 16713373. Quoted by Kuhn et al. (2012).
- ^ Kuhn, Jens H.; et al. (2012). "Virus nomenclature below the species level: a standardized nomenclature for natural variants of viruses assigned to the family Filoviridae". Archives of Virology. 158 (1): 301–311. doi:10.1007/s00705-012-1454-0. ISSN 0304-8608. PMC 3535543. PMID 23001720. Cited by User:Pol098 at Talk:Variant of Concern 202012/01 § Please do not call it a strain. (variant, mutation, change)
- ^ As of late December 2020, the only dialogues on Wikipedia about variant vs. strain are at Talk:Variant of Concern 202012/01 § Please do not call it a strain. (variant, mutation, change) and Talk:Cluster 5 § No category for strains? (as per a search for "variant" "strain").
D614G
Shouldn't even D614G be discussed here? — Preceding unsigned comment added by 83.185.44.225 (talk) 00:00, 1 January 2021 (UTC)
- I've added a section on D614G, sourced to two review articles. Fences&Windows 20:51, 2 January 2021 (UTC)
Naming and lineages
Biscuit-in-Chief, you write in your es, "→B.1.1.7: rename section to the official name, "Variant of Concern 202012/01". "B.1.1.7" is just a name use by Rambaut et al. in their system og lineages (see ref 7 and 8 as of 2021/01/01))". That's not quite right -- that the UK government using one name doesn't make it somehow more "official" in the global conversation. As for "just a name," well, yes, all names are "just" names—that doesn't convincingly argue for using or not using such names. More significantly, another entry in the same list uses Rambaut's lineage system (B.1.207), so it's weird to use more naming systems here than we have to, and doesn't promote comparability. I don't think highlighting the VOI/VOC numbering here is the most useful, because we're trying to compare variants to each other, not to rank them by the level of public health concern, either globally or specifically in the UK. jhawkinson (talk) 16:31, 4 January 2021 (UTC)
- Side note: Nexstrain amended their naming strategy 12 days ago, laying out their arguments in a blog post. Ain92 (talk) 13:43, 18 January 2021 (UTC)
Japan/Brazil variant
Does anyone have information on this variant? — Preceding unsigned comment added by 184.147.106.95 (talk) 22:49, 10 January 2021 (UTC)
- There's now a dedicated page, Lineage B.1.1.248, for this variant. SpookiePuppy (talk) 01:01, 23 January 2021 (UTC)
B1351 (South African variant)
Seems notable.[1] Zazpot (talk) 20:48, 12 January 2021 (UTC)
- This is another name for 501.V2 variant which is already included. ArcMachaon (talk) 21:43, 12 January 2021 (UTC)
References
- ^ Sample, Ian (2021-01-12). "Public Health England steps up surveillance of South African Covid variant". the Guardian. Retrieved 2021-01-12.
'A701B'(?)
Little seems to be forthcoming re mutation 'A701B'(sic) beyond the two existing refs from The Straits Times, 23 December 2020 and GMA News, 27 December 2020. However, I note that there is a more technical source (mirrored on the FB page for Noor Hisham Abdullah, who is quoted as the source for the Straits Times article) which refers to 'A701V' in very similar terms.
- On 25 December 2020, the organisation 'Kementerian Kesihatan Malaysia/ covid-19 Malaysia' wrote: "A701V mutation; ... was first found among 22 SARS-CoV-2 sequences from the Benteng LD clusters and subsequently passed on to the vast majority of third wave clusters in Peninsular Malaysia and Sabah."[1]
cf.
- The Straits Times (currently ref 46) wrote: " Malaysia's Health Ministry said on Wednesday (Dec 23) it has identified a new Covid-19 strain in the country from samples taken in Sabah. The ministry's director-general, Tan Sri Dr Noor Hisham Abdullah, said it is still unknown whether the strain - dubbed the "A701B" mutation - is more infectious than usual. "It is similar to a strain found in South Africa, Australia and the Netherlands," he said at his daily news conference."[2]
- GMA News (currently ref 47) wrote: "reports that a new coronavirus disease 2019 (COVID-19) variant has been detected in neighboring Sabah, Malaysia... Malaysia’s Health Ministry has dubbed the new variant as the "A701B" mutation"[3]
It seems to me that we have the same thing under discussion, but with its name potentially misreported (possibly via a mis-hearing?)
I am therefore minded to change the existing section "A701B" to "A701V" retaining A701B in the text but appending '(sic)', and add the following:
- On 25 December 2020, the organisation 'Kementerian Kesihatan Malaysia/ covid-19 Malaysia' described a mutation A701V as circulating and present in 85% of cases (D614G was present in 100% of cases) in Malaysia.[1]
- This (A701V) mutation has the amino acid Alanine substituted by Valine at codon position 701 in the spike protein. The source states that globally, South Africa, Australia, Netherlands and England also reported A701V at about the same time as Malaysia.[1] In GISAID, the prevalence of this mutation is found to be about 0.18%. of cases.[1]
Yadsalohcin (talk) 19:45, 15 January 2021 (UTC)
- I've removed this mutation from the summary of variants until further information can be found. No information about a specific variant has been found and there are conflicting reports about the mutation itself. Side note: For those that are unaware, A701B indicated an alanine (GCN) to aspartic acid (GAY) or asparagine (AAY) mutation, whereas, as you have written, A701V indicates alanine (GCN) to valine (GUN). ArcMachaon (talk) 02:40, 16 January 2021 (UTC)
- And I've commented the A701V. I think we should wait till this gets clarified by an RS. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 03:41, 16 January 2021 (UTC)
References
- ^ a b c d The current situation and Information on the Spike protein mutation of Covid-19 in Malaysia 25 December 2020 Kementerian Kesihatan Malaysia/ covid-19 Malaysia/ covid-19.moh.gov.my accessed 15 January 2021
- ^ "Malaysia identifies new Covid-19 strain, similar to one found in 3 other countries". The Straits Times. 23 December 2020. Retrieved 10 January 2021.
- ^ "Duterte says Sulu seeking help after new COVID-19 variant detected in nearby Sabah, Malaysia". GMA News. 27 December 2020. Retrieved 10 January 2021.
New variant P.1
There's a new variant P.1 (descendent of B.1.1.28) that's getting some attention and may reach the level of notability to deserve a section in this article.
- @billhanage (Jan 12, 2021). "There is a new 'variant' clearly identified today, P.1. And it is worth saying a little about what we have learned about it, and from the variants we have identified so far https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 1/quite_a_few" (Tweet) – via Twitter.
citing
- Nuno R. Faria (Jan 12, 2021). "Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings". Virological.
(It may be what is referred to above as Talk:Variants of SARS-CoV-2/Archive 1#Japan/Brazil variant, but maybe not?) jhawkinson (talk) 14:04, 13 January 2021 (UTC)
- It does definitely seem of interest considering the RBD mutations, but I don't think there has has been enough reported of it to introduce it yet. I suspect there will be very soon, though. Regarding your last sentence, they're the same. https://cov-lineages.org/lineages/lineage_B.1.1.28.html says:
[P.1 is the alias] of B.1.1.28.1, Brazilian lineage with a number of spike mutaions with likely functional significance E484K, K417T, and N501Y. Described in https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586
. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 15:18, 13 January 2021 (UTC)- That reminds me that perhaps https://cov-lineages.org should be an EL on this page. jhawkinson (talk) 15:36, 13 January 2021 (UTC)
- I suspect there is some confusion going on both on this page and on Lineage B.1.1.248. Specifically, it appears there are 2 widely discussed variants thought to be originated in Brazil. One is referred to as P.1, B.1.1.28.1, and 20J/501Y.V3. It has both N501Y and E484K mutations. Another is referred to as P.2, B.1.1.248, and 20B/S.484K. According to "COG-UK report on SARS-CoV-2 Spike mutations of interest in the UK 15th January 2021" (PDF). P.2 has E484K but not N501Y. Both this article and Lineage B.1.1.248 seem to combine information about P.1 and P.2 assuming it is the same lineage. However, "National Institute of Infectious Diseases report "New Variant Strainof SARS-CoV-2Identified in Travelers from Brazil"" (PDF). seems to use the name B.1.1.248 for P.1 instead of P.2 adding to the confusion. I'm trying to verify the above claims to resolve the confusion. Vikasatkin (talk) 18:13, 16 January 2021 (UTC)
- @Vikasatkin: I regret that, as regards 'Variants_of_SARS-CoV-2 / $ B.1.1.248', lack of clarity is largely my fault. To be explicit, the first sentence of the second para of Lineage B.1.1.248 could perhaps usefully have read
- "A preprint of a paper by Carolina M Voloch et al. identified a novel lineage of SARS-CoV-2 in circulation in Brazil, 'B.1.1.248', which originated from B.1.1.28." -and similarly, later in that para, 'novel variant lineage' could have '(B.1.1.248)' added to remove all possibility of doubt. I must re-read the source for further distinctions...
- Use of P1 and P2 is perhaps best kept back for more detail in the main article. Yadsalohcin (talk) 22:20, 16 January 2021 (UTC)
COH.20G/501Y
Researchers Discover New Variant of COVID-19 Virus in Columbus, Ohio I don't know enough to be able to tell if this is already generally covered in the article, specifically regarding the "20G" ID. Mapsax (talk) 00:23, 16 January 2021 (UTC)
- Hi Mapsax, thanks for this, for the moment the 'obvious place' seems to me to be a further addition to the sub section Notable mutations/ N501Y... only time will tell what we should eventually do with this information. Yadsalohcin (talk) 01:07, 16 January 2021 (UTC)
Other variants in the U.S. and Canada
At the risk of Ohio becoming WP:UNDUE, I wanted to mention that I've heard of other variants in the mainstream media throughout North America – as I type this, they've been reported in New York, Louisiana, Texas, and Ontario – but I don't know for sure if those are existing variants (I do know that reports call the variant in California the "UK variant") and wouldn't know which sources would be accepted as RS in this context. Mapsax (talk) 23:39, 16 January 2021 (UTC)
Nonspecific incoming redirects
There are a bunch of non-specific incoming redirects. They need to be handled with hatnotes.
{{redirect|Coronavirus variant|variations of coronavirus|Coronavirus}}{{redirect|New coronavirus variant|recent variations of coronavirus|Novel coronavirus}}{{redirect|New Covid|COVID nomenclature|wikt:COVID{{!}}Wiktionary]]|recently identified coronavirus diseases|Coronavirus disease}}
Clearly, this article doses not cover all coronaviruses, so it should carry hatnotes to handle these incredibly generic terms.
-- 70.31.205.108 (talk) 14:43, 18 January 2021 (UTC)
"COVID-19 variant"
Various incoming redirects like COVID-19 variant point here, shouldn't they point to coronavirus disease 2019? The various different presentations of the disease are covered in the main disease article.
If they remain pointed here, there needs to be a hatnote for it, like:
{{redirect|COVID-19 variant|variations in the course of COVID-19 disease progression|Coronavirus disease 2019}}
-- 70.31.205.108 (talk) 14:53, 18 January 2021 (UTC)
- You can do that if you want to, but "COVID-19 variant" should redirect to this article: even though technically incorrect, people who don't know much about virology might type in "COVID-19 variant", expecting something like a list of variants of SARS-CoV-2. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:58, 19 January 2021 (UTC)
Nomenclature table
The nomenclature table is a bit of a mess. That's not really our fault: the nomenclature in general is "a bloody mess at the moment", as one scientist put it.[2] To avoid further confusion—and because we've already nosedived into WP:OR territory—I suggest deleting the table for now until a secondary source compiles the nomenclatures, like Alm et al. did (although that table, of course, is outdated). —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:01, 20 January 2021 (UTC)
- Hello there, B-i-C, yes, the nomenclature table is undoubtedly a bit of a mess, for the reason stated, and because virusses in their evolutionary development don't necessarily choose to accommodate themselves to the layout on a table. However, with one exception (see below) it verifiably follows the reliable sources available to us at the moment and I would contend is a potentially helpful summary of variant names (and with 3 different schemes available there's plenty of room for getting lost).
- As a demonstration of its worthiness, it for example draws attention to the fact that I had over eagerly added CAL.20C to it when in fact we have so far only two news media sources for CAL.20C on this page, which (I think, on closer inspection) do not define its position within any of the naming schemes- its name in this sense would lead one to assume (sorry!) that it belonged on the 20C line, whereas the 'Nextstrain-amendation' source mentions 20G: (derived from 20C... 'concentrated in the United States') which might suggest that it should be with 20G, so, time to cancel said speculation and remove CAL.20C from the table. The table is then all fully supported once more by the refs. It will be interesting to see what more we learn about CAL.20C in the coming weeks. Yadsalohcin (talk) 15:19, 20 January 2021 (UTC)
- Thank you Yadsalohcin! I was going to add it to the table as well but since the Cedars-Sinai preprint is mythological at this point and there are no other references in the literature, I held off. "Interesting" is one way to put it... Mar2194 (talk) 16:31, 20 January 2021 (UTC)
Linkouts to resources
Human genes and protein pages link to various interactive viewers where one can see data about the gene graphically, on the chromosome, 3D structures, protein data etc. For Covid variants there is currently no linkout on this page. I work for one of these websites (UCSC Genome Browser) and I'm wondering if I should start linking to the info graphics that we generate, with mutation frequency, where they are in the SARS-CoV-2 genome, what the protein models on this mutation are, etc. One place could be the nomenclature table. There are a few other resources this page could link to, Nextstrain for example, they may have a URL scheme where the viewer zooms directly onto a mutation and one can get the map of where it currently is found. Maximilianh (talk) 09:35, 21 January 2021 (UTC)
- Hi Maximilianh, going beyond this basic table which cross-references nomenclatures, there are several obvious possible extras- in particular:
- date and location first detected,
- date of sample set (as there are often delays testing and reporting) and region of 'current' greatest prevalence
* principal mutations (but this is now taken care-of in the table in the summary section)- -Further to these, we have a number of refs and a footnote link from the table, so I don't see why we shouldn't also include refs linking to various external viewers and map pages... seems a potentially valuable hub for information- I'd love to see them here. Yadsalohcin (talk) 00:55, 22 January 2021 (UTC)
Fresh preprint about South African variant and antibody escape
Do this worth mentioning in article? --Unxed (talk) 18:52, 21 January 2021 (UTC)
- Not sure. It's a preprint, so we might wanna wait till peer-reviewed version is available. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 20:03, 21 January 2021 (UTC)
- We mention a couple of Preprints already; I would have thought that provided its 'preprint nature' is clearly stated, mention of the likely upshot of this study would seem in order (tho' maybe in the 501.V2 variant main article first?) Yadsalohcin (talk) 01:00, 22 January 2021 (UTC)
- Added link to 501.V2 talks page, so it could be discussed there also --Unxed (talk) 14:18, 22 January 2021 (UTC)
- We mention a couple of Preprints already; I would have thought that provided its 'preprint nature' is clearly stated, mention of the likely upshot of this study would seem in order (tho' maybe in the 501.V2 variant main article first?) Yadsalohcin (talk) 01:00, 22 January 2021 (UTC)
WIV04/2019
Shouldn't the "original" strain(s) also have a section? (the two major variants identified in Wuhan in January) -- 70.31.205.108 (talk) 14:47, 18 January 2021 (UTC)
- The original strain is covered at Severe acute respiratory syndrome coronavirus 2. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:58, 19 January 2021 (UTC)
- This article is "variants of", so it would seem logical to have a section for that (the original variants), with a short summary, and a redirect to the base article -- 70.31.205.108 (talk) 08:04, 21 January 2021 (UTC)
- A sequence zero/reference sequence cannot be a variant as it is not different from the reference sequence, as per PMC 3535543:
Van Regenmortel defined a virus variant as an isolate or a set of isolates whose genomic (consensus) sequence(s) differ(s) from that of a reference virus.
—Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 08:11, 21 January 2021 (UTC)- The reference sequence is itself a variant in the spectrum of all variations. Though not such within the field's scientific literature, it is such in as a logical consequence, since the original virus is never the reference sequence, as the sequence that is referenced is inevitably a descendant of the original, sometimes a much later one. -- 70.31.205.108 (talk) 03:50, 25 January 2021 (UTC)
- A sequence zero/reference sequence cannot be a variant as it is not different from the reference sequence, as per PMC 3535543:
- This article is "variants of", so it would seem logical to have a section for that (the original variants), with a short summary, and a redirect to the base article -- 70.31.205.108 (talk) 08:04, 21 January 2021 (UTC)
Article's content does not meet its title
The title of the article is "Variants of SARS-CoV-2". Yet the avowed content is "[those] that are or have been believed to be of particular importance". This article needs an expert to explain the criteria for importance and why unimportant variants can be omitted. It would be useful for that expert to explain whether the rate of all new variants is consistent with that of other viruses or whether the preventive measures we have adopted are part of an arms race with the virus. Robert P. O'Shea (talk) 05:48, 23 January 2021 (UTC)
- I do not entirely disagree, but it's only to be expected that an article about "Variants of X" talks about variants of X in general, and then discusses notable variants of X further. Establishing strict criteria for notability, other than wikipedia's general and rather fluffy criteria, is probably not possible at this point. I think the lead is sufficiently clear about this, saying "some are or have been believed to be of particular importance".--Nø (talk) 12:15, 23 January 2021 (UTC)
- From https://www.who.int/bulletin/volumes/98/7/20-253591/en/ (used in the article): "We detected in total 65776 variants with 5775 distinct variants". Considering these data are from at least May 2020, maybe even February, and that they obviously only looked at a limited number of genomes, it is only reasonable to suggest that there are millions of variants. We can't write about them all. We write about variants that have been reported widely in media (whether that be popular or specialised) and are determined to be of importance in one way or another by these sources. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 18:06, 27 January 2021 (UTC)
E484K
Is this RS? https://www.biorxiv.org/content/10.1101/2020.12.31.425021v1.article-info Charles Juvon (talk) 21:32, 24 January 2021 (UTC)
- Also, see: https://science.sciencemag.org/content/371/6527/329 Charles Juvon (talk) 22:20, 24 January 2021 (UTC)
Remove the ticks and crosses
Please remove the {{ya}} and {{na}} from the summary table. A tick usually means yes, and the x cross usually means no, and it took me some time before realising that this is not what is meant in the article. For example under transmissibility I would read it as yes for the tick (therefore transmissible), and no for x (therefore not transmissible), which is exactly the opposite of what is meant there. Using it this way only leads to confusion. The colours alone are enough to indicate good or bad outcome. Hzh (talk) 23:48, 1 February 2021 (UTC)
- I agree, I initially created the table in the inverse - where ticks mean yes and crosses mean no. I understand that the red and green colours may have caused some confusion, so I have edited the table to remove the symbols altogether as you suggested. ArcMachaon (talk) 15:43, 2 February 2021 (UTC)
Roman numerals in headings
Roman numerals, or any other number should not be used in the headings for several reasons:
- The headings are already numbered in the contents box and do not need to be renumbered
- The numbering system may be mistaken for something relevant, such as an official designation or numbering system
- It is unwieldy and not particularly aesthetic
- It goes against WP:HEADING, which states headings should not be numbered etc.
Therefore, I have removed the numbering from the article. ArcMachaon (talk) 02:45, 16 January 2021 (UTC)
- thanks for post--Ozzie10aaaa (talk) 20:11, 15 February 2021 (UTC)
Summary table
Currently, without the four pipes seperating "Date" and "Transmissibility", I am seeing the summary table like this: https://imgur.com/a/ddI6Ot6 (notice position of "Transmissibility", "Virulence"; and "Antigenicity"), both on my desktop and laptop. I realise some people might be seeing it correctly (I also see it correctly in the mobile version). Anyone know how to fix this? —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:38, 16 January 2021 (UTC)
- thank you--Ozzie10aaaa (talk) 20:11, 15 February 2021 (UTC)
Seven U.S. variants
Emergence in late 2020 of multiple lineages of SARS-CoV-2 Spike protein variants affecting amino acid position 677 (not peer reviewed as of now) This study is showing up in a number of mainstream media reports. Mapsax (talk) 22:59, 15 February 2021 (UTC)
- yes Ive seen it as well--Ozzie10aaaa (talk) 20:56, 16 February 2021 (UTC)
Table Colors
Could we swap red and green background color in the table for a more intuitive representation like "red=danger, green=irrelevant" ? --jOERG 17:20, 28 January 2021 (UTC) — Preceding unsigned comment added by Joerg rw (talk • contribs) done --jOERG 18:48, 28 January 2021 (UTC) — Preceding unsigned comment added by Joerg rw (talk • contribs)
Cluster 5 not extinct?
German news n-tv reports that one dead in Bavaria from a strain with a Cluster 5-like mutation. It's just on their Covid ticker, but there should be some official or other usable RS; the source of the claim is not given more specifically than "Bavarian labs". 213.182.112.25 (talk) 22:41, 14 February 2021 (UTC)
- You say cluster 5–like mutation; does this mean only Y453F? Because in that case, it wouldn't be cluster 5 since there aren't necessarily the several other defining mutations. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:57, 14 February 2021 (UTC)
- A team from the SSI confirmed that Cluster 5 was not in circulation in the human population as of 1 February 2021 see here, section "Cluster 5 variant". I have added this to the section and removed the banner. Biscuit-in-Chief is likely correct in their assessment of the headline. If more information about the prevalence of specific mutations is found, feel free to add it to the "notable mutations" section. ArcMachaon (talk) 20:46, 18 February 2021 (UTC)
- The news item had no more details than "the strain found in Danisk mink farms" or similar. Thanks for the update, but it is not relevant to that case - the Feb 1 SSI paper is probably correct in Cluster 5 being extinct in Denmark (it would have been found since otherwise, because the Danes were actively looking for it), but the appearance in Bavaria was after Feb 1 (it was probably an infection that occurred locally at the end of January and no later than Feb 2, assuming the usual German testing/reporting workflow - the infected person was probably symptomatic as Germany does not routinely test people without symptoms, and add to this 2-4 days delay for reporting the original PCR test result, and another ~3 days before the sequencing results are reported), and how it would have gotten there is entirely mysterious. Given that Germany is rather lacklustre in its sequencing effort, I suspect that it's some Cluster-5-like convergent mutation, and that they confused it because only a few snippets are sequenced (Germany does apparently not distinguish between Lineage B.1.351 and B.1.525, or VOC-202012/01 and VOC-202102/02; tho both strains with additional E484K are reasonable to expect there given their spread elsewhere in Europe, none have been reported to date). I will try and find any follow-up to the Bavarian report, which is puzzling and if true kinda disturbing. 213.182.113.251 (talk) 13:08, 19 February 2021 (UTC)
- Ah yes, that was easy:
- https://www.swp.de/panorama/corona-mutation-bayern-neu-ulm-ansbach-daenemark-coronavirus-bayreuth-verbreitet-sich-b117-b1351-p1-zahlen-54700406.html
- https://www.stimme.de/suedwesten/nachrichten/pl/labor-entdeckt-daenische-corona-mutation-in-bayern;art19070,4443059
- https://www.merkur.de/welt/coronavirus-deutschland-bayern-mutation-nerz-cluster-5-neuulm-variante-tier-drosten-rki-90203580.html
- 10 cases with at least 1 fatality reported as of Feb 16; the first case reported Jan 27 already, meaning infection most likely occurred about Jan 10. 2 sequencing labs found it, one Austria (1 case), the other in Germany (the other cases), so it's likely to be genuine - though details are still lacking, it's just "Cluster 5" or "the Danish mink strain". Cases cluster in Neu-Ulm county, but at least one in Ansbach which is quite some way distant.
- The most consistent hypothesis is that someone on holiday or business travel picked it up in the very days before it was eradicated in Denmark. It's highly unlikely that more than 1, maybe 2 people imported the strain to Bavaria (probably Neu-Ulm area), given the distance involved; any spread beyond their household probably only happened during the Christmas/New Year week. That would result in the half-dozen or so chains of transmission known to be active at the end of January.
- I've put the links into the article (these are all local/regional daily newspapers, none of them "yellow press", and hence qualify as RS as long as no proper scientific source is available), but you may wanna tidy it up. I have copypasted it to the main Cluster 5 article too, but again, only quick-and-dirty. It should be OK for the meantime, but any tidying up (linking the sources; at least Münchner Merkur has an article) is also appreciated.)
- I hope there's a scientific study soon, because this is very puzzling (though not entirely implausible; it would still be awfully bad luck - literally "the one that got away") and worrying (Bavaria does a really bad job as regards containment; they neglect fighting domestic transmission in favour of closing external borders). At least they do sequence, though not yet (but hopefully soon) in an organized fashion. 213.182.113.251 (talk) 14:05, 19 February 2021 (UTC)
- @213.182.113.25: I have reverted your edits—the popular-media reports keep using to cluster 5 and "the Danish mink variant" interchangeably, which clearly shows they haven't understood it correctly: the SSI, as explained in this report, found five clusters of mink variants as of early November 2020; cluster 5 was just the concerning one. We should wait with reporting on this till there are reliable medical sources from subject-matter experts we can cite. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 15:21, 19 February 2021 (UTC)
- The news item had no more details than "the strain found in Danisk mink farms" or similar. Thanks for the update, but it is not relevant to that case - the Feb 1 SSI paper is probably correct in Cluster 5 being extinct in Denmark (it would have been found since otherwise, because the Danes were actively looking for it), but the appearance in Bavaria was after Feb 1 (it was probably an infection that occurred locally at the end of January and no later than Feb 2, assuming the usual German testing/reporting workflow - the infected person was probably symptomatic as Germany does not routinely test people without symptoms, and add to this 2-4 days delay for reporting the original PCR test result, and another ~3 days before the sequencing results are reported), and how it would have gotten there is entirely mysterious. Given that Germany is rather lacklustre in its sequencing effort, I suspect that it's some Cluster-5-like convergent mutation, and that they confused it because only a few snippets are sequenced (Germany does apparently not distinguish between Lineage B.1.351 and B.1.525, or VOC-202012/01 and VOC-202102/02; tho both strains with additional E484K are reasonable to expect there given their spread elsewhere in Europe, none have been reported to date). I will try and find any follow-up to the Bavarian report, which is puzzling and if true kinda disturbing. 213.182.113.251 (talk) 13:08, 19 February 2021 (UTC)
- The previously linked swp.de source says: "Am Mittwochabend ließ das Landratsamt Neu-Ulm jedoch vermelden, dass ihnen Informationen über einen gesicherten Nachweis der dänischen Variante in Proben aus Bayern bislang nicht vorliegen" (translated: "Wednesday the Neu-Ulm District office said that they did not have information about reliable evidence showing the Danish variant in Bavaria"). Additionally, the sources keep on randomly going between "Cluster 5", "Danish variant/Danish mink variant" and "mink variant". These are not the same thing. As mentioned by Biscuit-in-Chief, cluster 5 is a very specific mink variant (hence its name, "5"), which only ever was found in 12 people in/near one village several months ago. A number of other mink-related variants do exist and they have been involved in more cases in a wider region, but they're not known to be particularly problematic; their behavior is similar to "ordinary" COVID-19. All mink-related in Denmark have, collectively, been referred to as the "Danish (mink-related) variants"; all these are now extinct in the country with no recent detections. Then there are mink-related variants without further specification, which have been discovered in several countries with outbreak in mink farms. They're generally defined by the Y453F mutation in combination with others, and part of several different clades (i.e., they emerged independently of each other). However, the problem is that the Y453F mutation, while usually linked to mink, also can occur under other circumstances, as shown by its developent in an immunocompromised, long covid patient (Δ69-70 itself is quite common, having been detected in at least six independent clades, with first detection more than a year ago in Thailand and Germany). I've considered updating/correcting the Cluster 5 article and will probably do it one of the next days, including its "Implications for human health" section which confuses cluster 5 (=12 cases) and "Danish (mink-related) variants" (=more cases); this is also described in the recently published review. However, until we get clear information about the German cases, it is impossible to say if or where they belong in this, but I'd be very surprised if they're real cluster 5. They're in all probability "Danish (mink-related) variant" or some other variant with a Y453F mutation. I've just checked GISAID and Nextstrain and they have no cluster 5/Danish (mink-related) variant/Y453F mutation from Germany. Finally, I am very puzzled by the claim in the most recent German article that these have been little studied; there have been a good handful of scientific articles about them. RN1970 (talk) 00:24, 20 February 2021 (UTC)
Garmisch Variant turns out to be B.1.1.134, not novel
Turns out the unusual COVID variant that was detected in Garmisch-Patenkirchen are not novel, but belong to the sporadically, previously seen B.1.1.134 lineage. Unfortunately, I only have this German source on hand: https://www.br.de/nachrichten/bayern/corona-variante-aus-garmisch-partenkirchen-ist-entschluesselt,SMjr7L1 134.3.255.250 (talk) 13:46, 31 January 2021 (UTC)
- thank you for post--Ozzie10aaaa (talk) 13:25, 22 February 2021 (UTC)
L452R in California
I have been seeing multiple articles about L452R variant in Santa Clara County, CA. News article 1, News article 2,County press release. Should this get a section on this page? --Gimelthedog (talk) 21:30, 18 January 2021 (UTC)
- I say we wait a bit. There seems to quite a bit on L452R (it's mentioned in Cell as a mutation that is
markedly resistant to some mAbs
), [1] but it's only one mutation and has been around since at least May of last year[2]—maybe even February (per "Methods" on p.1). Furthermore, so far all reports have been on transmission with L452R in California; I personally think we should wait for it spread to other countries before writing about it because of the risk of becoming WP:UNDUE. —Happy New Year from Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 14:58, 19 January 2021 (UTC)
- Also referred to in the New York Times as CAL.20C[3]; there's little reason to wait before adding a variant, people will come here to look for it — it's definitely a variant and that should be enough for notability. The level of information and its import will of course change over time, and we'll change the article appropriately. jhawkinson (talk) 23:18, 19 January 2021 (UTC)
- Sorry I didn't check the talk page before adding the CAL.20C variant section... I tend to agree with jhawkinson in that there's no real reason to wait before adding this variant to the page. I am a little annoyed with the Cedars-Sinai team however... In their press release, they claimed that there is a preprint on MedRxiv.org [4] but I can't seem to find any 2021 submissions by any of the coauthors listed in the press release. Mar2194 (talk) 00:40, 20 January 2021 (UTC)
- The claim in the press release is that the preprint was "submitted" to medRxiv. You might need to wait a bit before it gets approved by medRxiv and made public. Could be longer if medRxiv rejects the first submission. In any case, I don't think Wikipedia allows medRxiv references. Also, the URL for the press release has gone bad, but I found it cached on Google. Jaredroach (talk) 03:56, 25 February 2021 (UTC)
- Sorry I didn't check the talk page before adding the CAL.20C variant section... I tend to agree with jhawkinson in that there's no real reason to wait before adding this variant to the page. I am a little annoyed with the Cedars-Sinai team however... In their press release, they claimed that there is a preprint on MedRxiv.org [4] but I can't seem to find any 2021 submissions by any of the coauthors listed in the press release. Mar2194 (talk) 00:40, 20 January 2021 (UTC)
References
- ^ https://www.cell.com/cell/pdf/S0092-8674%2820%2930877-1.pdf, p.1
- ^ https://www.who.int/bulletin/volumes/98/7/20-253591/en/, figure 3. It's hard to spot, but you can go to doi:10.2471/BLT.20.253591 and use CTRL + F to find it along the second row of figure 3 on p. 6.
- ^ Carl Zimmer (January 19, 2021). "New California Variant May Be Driving Virus Surge There, Study Suggests". Event occurs at 5:24 p.m. ET.
Researchers found that the variant originated in California and showed up in more than half of samples tested last week by researchers in Los Angeles.
- ^ https://www.cedars-sinai.org/newsroom/local-covid-19-strain-found-in-over-one-third-of-los-angeles-patients/
Spanish variant
In October 2020, a Spanish variant of COVID-19 was discovered, and known as 20A.EU1. This is what caused the second wave of infections in the UK and other European countries following the "travel corridors" idea. The article stated that the discovery had not been peer-reviewed so this is probably why this wasn't given a mention here... though I wonder if it could this time now that I've brought it up. What does everyone think? Bryn89 (talk) 18:12, 21 February 2021 (UTC)
- This is also known as B.1.177 and appears in the table as a clade. Though it contains the A222V mutation, it doesn't seem to affect transmissibility - it was just the strain that holidaymakers happened to bring back from Spain at the end of summer (see Sci Am and BBC Future. If you can find review articles discussing it then a brief section could be added. One says "Prior to B.1.1.7, B.1.177 (also called “20A.EU1”) was the dominant variant in the UK, although it is not believed to have arisen due to increased transmissibility. Since the earliest sequenced sample of B.1.1.7 (September 20, 2020), it started to displace B.1.177 and other variants in the UK."[3] Fences&Windows 21:42, 25 February 2021 (UTC)
- I do think that this variant received enough coverage in media and medical sources to fulfil the notability criteria. It is also notable because it became a main lineage and is still the dominant strain in parts of Europe. I also note it is mentioned on the SARS-CoV-2 main page but not here:
In July 2020, scientists reported that a more infectious SARS-CoV-2 variant with spike protein variant G614 has replaced D614 as the dominant form in the pandemic. In October 2020 scientists reported in a preprint that a variant, 20A.EU1, was first observed in Spain in early summer and has become the most frequent variant in multiple European countries. They also illustrate the emergence and spread of other frequent clusters of sequences using Nextstrain.
- Overall it would probably be a good WP:BOLD addition to this page. ArcMachaon (talk) 14:49, 26 February 2021 (UTC)
Vandalism / Spamming
ZRS2012 seems to be repeatedly vandalizing this page, and has done similar things to other covid related pages. Is there any recourse? TimeEngineer (talk) 00:50, 18 February 2021 (UTC)
- Ive got this article on my watchlist--Ozzie10aaaa (talk) 02:27, 18 February 2021 (UTC)
- Surely the user:ZRS2012 must be in violation, saying he/she is the same as recently blocked user:Theusernameistaken, who indeed as been blocked indefinitely. 178.155.171.181 (talk) 19:35, 18 February 2021 (UTC)
- On this basis, how many warnings are required on the user:ZRS2012 talk page before blocking further edits from them can be requested? This seems a somewhat aggravated case -see WP:AIV and the warning templates - I see that Biscuit-in-Chief has already posted a 'vandalism3' ("please stop") notice at User talk:ZRS2012 -Hope this helps, Yadsalohcin (talk) 23:28, 18 February 2021 (UTC)
- Yeah, we can probably request that now. I posted vandalism3 before their most recent vandalism. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:42, 18 February 2021 (UTC)
- Actually, we most definitely can, per WP:EVADE. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:44, 18 February 2021 (UTC)
- Yeah, we can probably request that now. I posted vandalism3 before their most recent vandalism. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:42, 18 February 2021 (UTC)
- On this basis, how many warnings are required on the user:ZRS2012 talk page before blocking further edits from them can be requested? This seems a somewhat aggravated case -see WP:AIV and the warning templates - I see that Biscuit-in-Chief has already posted a 'vandalism3' ("please stop") notice at User talk:ZRS2012 -Hope this helps, Yadsalohcin (talk) 23:28, 18 February 2021 (UTC)
- Surely the user:ZRS2012 must be in violation, saying he/she is the same as recently blocked user:Theusernameistaken, who indeed as been blocked indefinitely. 178.155.171.181 (talk) 19:35, 18 February 2021 (UTC)
@Yadsalohcin and TimeEngineer: I've reported them at Wikipedia:Administrators'_noticeboard/Incidents#ZRS2012_evading_block_and_doing_disruptive_edits. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 23:55, 18 February 2021 (UTC)
- They're now blocked indefinitely. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 00:17, 19 February 2021 (UTC)
@Biscuit-in-Chief: They seem to be back as "GaatWild". Same sentence structure in the comments and similar revisions TimeEngineer (talk) 21:06, 23 February 2021 (UTC)
- @TimeEngineer: WP:SPI. —Biscuit-in-Chief :-) (/tɔːk/ – /ˈkɒntɹɪbs/) 21:26, 23 February 2021 (UTC)
- @Biscuit-in-Chief: done here link TimeEngineer (talk) 14:35, 25 February 2021 (UTC)
@Ifnord:, @Biscuit-in-Chief: and @TimeEngineer: we seem to have a resumption of this disruptive vandalism, previously by User:ZRS2012 and User talk:GaatWild, this time from User:DORKKLOK. I see Ifnord has placed a warning template on their user page (thanks) and I have now alerted the admin previously involved (LuK3 (Talk)). Yadsalohcin (talk) 22:00, 5 March 2021 (UTC)
Indian "double" mutant
It looks like another variant has been discovered in India: https://www.bbc.com/news/world-asia-india-56507988 — Preceding unsigned comment added by 2A01:799:B22:4F00:CD72:8884:932F:220 (talk) 19:26, 24 March 2021 (UTC)
- India reports 'double mutant' coronavirus variant as daily deaths reach year's high CBCyes interesting--Ozzie10aaaa (talk) 19:33, 24 March 2021 (UTC)
Proposal for shorthand redirect page...?
Hello, I'm sorry. I was curious if maybe a few pages for shorthand phrases like "COVID-19 VOC" could be created to redirect to this article since it seems natural to me that people would use such keywords to get to this page quickly and more often than typing out "variants of SARS-CoV-2" or something. I'd make the redirect of it myself but I actually don't know if I'm allowed to do so (especially as a guest user) or if I'd have to make a request to have it done instead. Thank you. 65.92.88.128 (talk) 22:25, 24 March 2021 (UTC)
- COVID variant already exists, although I suppose we could create "COVID-19 VOC" as well. JackFromReedsburg (talk | contribs) 22:31, 24 March 2021 (UTC)
New Variant ‘double mutant’ strain named B.1.617
Hi, I'm new to Wiki but I just came across the new variant B.1.617 and see it's not added. I don't know what I'm doing so I just though I would mention it. Thanks to everyone and their hard work here. — Preceding unsigned comment added by RegCliff (talk • contribs) 23:00, 8 April 2021 (UTC)
- thank you for link--Ozzie10aaaa (talk) 23:04, 8 April 2021 (UTC)
Fin-796H variant (B.1.1.318 Lineage)
Newly discovered Fin-796H variant is likely to have some historical significance as it includes a mutation which makes it harder to detect with some PCR-tests, bears similarities to both British and South African variants (E484K mutation included) and appears to be from a distinct lineage that has gone unnoticed for some time. It is unlikely to have originated in Finland where it was first discovered, as the local rate of infection is relatively low and this variant is said to contain at least 15 distinct mutations. Not much seems to be known yet about how widespread, contagious or vaccine-resistant it is, however it was recently sequenced and had it's sequence sent to GISAID. I'll be leaving inclusion decisions and wording choices up to someone more experienced... Sources (in English): https://yle.fi/uutiset/osasto/news/new_coronavirus_variant_discovered_in_finland/11796958 https://www.coronaheadsup.com/coronavirus/second-case-of-variant-fin-796h-confirmed-in-finland-four-more-fin796h-infections-likely/ Nuihc88 (talk) 07:21, 7 March 2021 (UTC)
- I think it was discovered some almost a month ago, so it isn't quite recent, but agree that it seems notable. What's actually recent is a French variant discovered a week ago which also fails on all three PCR gene targets. Ain92 (talk) 12:28, 24 March 2021 (UTC)
Looks like this variant is part of the B.1.1.318 lineage and most likely originated in Nigeria, but all the news reporting i could find about it seems shoddy and lacks primary sources. Nuihc88 (talk) 20:08, 19 April 2021 (UTC)
New variant B.1.618
Hi, can anyone well versed with the variant's notability, add about new variant B.1.618[1][2][3] that been supposed to be originated in India and has been found in US, Switzerland, Singapore and Finland as per reports.[4] Thank you. Run n Fly (talk) 14:38, 21 April 2021 (UTC)
References
- ^ Koshy, Jacob (21 April 2021). "New coronavirus lineage found in West Bengal". The Hindu.
- ^ Dasgupta, Binayak; Dutt, Anonna (21 April 2021). "New B.1.618 variants now among most sequenced". Hindustan Times.
- ^ Basu, Mohana (20 April 2021). "New Covid lineage B.1.618 identified from Bengal, 2nd in India after 'double mutant' virus". ThePrint.
- ^ Sharma, Milan (21 April 2021). "New immune escape coronavirus variant found in West Bengal, say experts". India Today.
New York Variant - Lineage B.1.526
Hello à tous,
I would like to inform you that a page is ready to be linked about the New York Variant: Lineage B.1.526. As I don't know how to insert it in this article, I have preferred to write a comment in the talk page, so that you guys who know more about the subject, would see how to deal with it, without adding in the See also section.
Thanks for all the work you do for the vulgarisation of this damned virus/disease/pandemic. --Anas1712 (talk) 23:58, 21 April 2021 (UTC)
 Partly done and anyone is welcomed to expand. Run n Fly (talk) 07:20, 22 April 2021 (UTC)
Partly done and anyone is welcomed to expand. Run n Fly (talk) 07:20, 22 April 2021 (UTC)
Thank you to editors
I thank all of the reasonable and hard-working editors who have been meticulously updating this extremely important article, which is well-structured and well-written, and its related companion articles. Acwilson9 (talk) 08:08, 24 April 2021 (UTC)
New variant A.VOI.V2 from Tanzania
South African scientists identified the most genetically divergent variant so far, see [4]. Ain92 (talk) 11:01, 27 March 2021 (UTC)
- "A novel variant of interest of SARS-CoV-2 with multiple spike mutations detected through travel surveillance in Africa". www.krisp.org.za. Retrieved 30 April 2021.--Ozzie10aaaa (talk) 22:39, 30 April 2021 (UTC)
New variant B1616
I have seen news articles about B1616 and that it can evade PCR tests. Shall we add em in? Hanami-Sakura (talk) 07:24, 12 May 2021 (UTC)
- Do you mean B.1.617? 'Evading' PCR sounds weird. Can you provide links. JuanTamad (talk) 07:48, 12 May 2021 (UTC)
https://www.medrxiv.org/content/10.1101/2021.05.05.21256690v1
https://www.ouest-france.fr/sante/virus/coronavirus/covid-19-la-direction-generale-de-la-sante-alerte-sur-un-nouveau-variant-detecte-en-bretagne-7187918 Unxed (talk) 13:33, 19 May 2021 (UTC)
Why do we still retain B.1.1.207 in the overview table?
Looks not so notable actually. Ain92 (talk) 19:46, 20 May 2021 (UTC)
- Agreed, and removed. --Fernando Trebien (talk) 01:48, 23 May 2021 (UTC)
Legend for Table Colors
Hello there, I see the table colors are unintuitive, it's not clear what the yellow and red and green are denoting. I would love someone could add a legend or otherwise visual assistant to explain the color coding Buumix (talk) Boring 13:45, 19 April 2021 (UTC)
- Done. --Fernando Trebien (talk) 02:39, 23 May 2021 (UTC)
Strains missing from overview table
The WHO "variants of interest"[5] now called Zeta, Theta, and Iota are missing from the Overview table. Given the WHO is worried about them, seems useful to include them. -- Beland (talk) 18:19, 31 May 2021 (UTC)
WHO names variants based on Greek alphabet?
Seems WHO has reported (31 May 2021) a new system of identifying Covid-19 variants - based on the Greek alphabet[1][2] - examples: the "UK variant" (B.1.1.7) is "Alpha"; the "South African variant" (B.1.351) is "Beta" - and so forth - perhaps this new system should be added to the main article? - in any case - Stay Safe and Healthy !! - Drbogdan (talk) 01:20, 1 June 2021 (UTC)
 Done - Brief followup - seems this has already been done - Stay Safe and Healthy !! - Drbogdan (talk) 02:00, 1 June 2021 (UTC)
Done - Brief followup - seems this has already been done - Stay Safe and Healthy !! - Drbogdan (talk) 02:00, 1 June 2021 (UTC)
References
- ^ Staff (31 May 2021). "SARS-CoV-2 Variants of Concern and Variants of Interest, updated 31 May 2021". World Health Organization. Retrieved 31 May 2021.
- ^ Howard, Jacqueline (31 May 2021). "WHO's new naming system for coronavirus variants uses Greek alphabet". CNN News. Retrieved 31 May 2021.
News (06/02/2021) worth considering?
Recent news (06/02/2021)[1] worth considering for the main article? - iac - Stay Safe and Healthy !! - Drbogdan (talk) 00:25, 3 June 2021 (UTC)
References
- ^ Speights, Keith (2 June 2021). "This Ominous Warning From Moderna Could Shake Up the COVID Vaccine Market - The pandemic isn't over yet -- even in the U.S." The Motley Fool. Retrieved 2 June 2021.
Pangolin origin information needs updating
This Though the emergence of SARS-CoV-2 may have resulted from recombination events between a bat SARS-like coronavirus and a pangolin coronavirus (through cross-species transmission)
needs update because more recent research downplays the possible role of pangolins for SARS-CoV-2 evolutionary history. See Severe Acute Respiratory Syndrome Coronavirus 2 for more current sources and up-to-date information. Forich (talk) 21:18, 4 June 2021 (UTC)
A Nature article by the Nextstrain team about a non-notable, yet quite interesting variant 20E(EU1)/B.1.177
[6]. I have no idea in what Wikipedia article it is best applicable, but found an overview Twitter thread to be interesting, take a look: https://twitter.com/firefoxx66/status/1401833676317593600 Ain92 (talk) 11:45, 7 June 2021 (UTC)
Dates in table
We have B.1.1.7 listed as first identified in February 2020, but it wasn't detected until December and was later found to have first been sequenced in September. The cited source, https://cov-lineages.org/lineages/lineage_B.1.1.7.html, does say February but it is clearly wrong: https://asm.org/Articles/2021/January/B-1-1-7-What-We-Know-About-the-Novel-SARS-CoV-2-Va ; https://www.sciencemag.org/news/2020/12/mutant-coronavirus-united-kingdom-sets-alarms-its-importance-remains-unclear. Do we need to stop using cov-lineages.org? Fences&Windows 11:31, 12 May 2021 (UTC)
- thank you for posting--Ozzie10aaaa (talk) 01:36, 24 May 2021 (UTC)
- February 2020 is the the date given by outbreak.info as well for any detection in the world. September 2020 is when it was first detected in the UK. --Fernando Trebien (talk) 18:03, 6 June 2021 (UTC)
- But I asked about this in the issue tracker of cov-lineages and they said that the lineages pages are not quality controlled and that there is a lot of data with incorrect labels. In cov-lineages, the best source for this information is the global report. It seems that outbreak.info also has no quality control either and it uses the same data source (GISAID). So it is better to rely on reviewed information provided by trusted sources such as the WHO, PHE, CDC, ECDC, ACI. --Fernando Trebien (talk) 11:29, 14 June 2021 (UTC)
Date of sequence of B.1.1.7
The article states that Lineage B.1.1.7 was sequenced in October 2020, sourced on a BBC article. I have this source[1] that says it was in late september. Which one is more accurate? Forich (talk) 20:05, 11 June 2021 (UTC)
- At the moment, your source is more accurate and the date given (20 September 2020) is exactly the same as given in the UK by cov-lineages and outbreak.info. The news piece referenced in the article was probably correct at the time it was published (December 2020) because it often takes time for new data to be integrated into the main databases. Note that this is not the oldest sequence in the world, which is February 2020, as reported in the Overview section. --Fernando Trebien (talk) 12:15, 13 June 2021 (UTC)
- Thanks, I believe it is updated now. Forich (talk) 20:43, 15 June 2021 (UTC)
References
- ^ Washington, Nicole L.; Gangavarapu, Karthik; Zeller, Mark; Bolze, Alexandre; Cirulli, Elizabeth T.; Schiabor Barrett, Kelly M.; et al. (May 2021). "Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States". Cell. 184 (10): 2587–2594.e7. doi:10.1016/j.cell.2021.03.052.
Use of {{risk}} in summary table
There seems to be some confusion with the colours in the summary table which seem subjective and not based upon a system or any reliable source. I would suggest that there are two options to fix this problem: return to the red-yellow-green (bad-pending-good) system previously used, or create a system a place it in comment form above the table for editors to see. For example:
<!--
Table Colours for clinical changes:
Risk should be "unknown" if risk is under investigation
Risk should be "low" if risk has been disproven by reliable investigation
Risk should be "medium" when "potentially", "likely", "indications of" etc. (NB. Reliable medical source still required)
Risk should be "high" if confirmed
Risk should be "very high" if critical and confirmed
Table Colours for spread:
Risk should be "very low" if thought extinct
Risk should be "low" if contained and localised
Risk should be "medium" if endemic to a specific region
Risk should be "high" if global spread
Risk should be "very high" if variant becomes a major lineage per reliable medical sources
-->
as one potential system. What are people's thoughts? ArcMachaon (talk) 23:53, 26 February 2021 (UTC)
- It is sensible to define better criteria. I'd seek some inspiration on the example risk matrix, though every risk matrix is different. For clinical changes, I'd consider risk as a combination of probability and harm level. When the harm level refers to an increase in expected deaths, I'd suggest basing the choices on the "critical" harm level with some adjustments, like this:
- "unknown" if risk is under investigation
- "verylow" if probability is described as rare or if risk has been disproven to a degree by reliable investigation or even "eliminated" if investigators are very confident of their results
- "low" if probability is described as unlikely
- "medium" if probability is described as possible or potential or as having some indications
- "high" if probability is described as likely
- "veryhigh" if probability is described as certain
- When risk refers to the harm level of disease leading lost work days but no deaths and no permanent disability, then I'd follow the "marginal" harm level. It is important to evaluate both probability and harm properly. For example, P.1. is currently described as "10–80% more lethal", but the link to CADDE says "can be 10–80% more lethal", meaning "potentially 10–80% more lethal". --Fernando Trebien (talk) 00:39, 8 March 2021 (UTC)
- Perhaps risk assessment framework from Public Health England can help refine the current criteria.[1] For example:
Changes Efficacy or neutralizing antibody activity Transmissibility Hospitalization / Mortality After natural infection From vaccination Monoclonal antibodies No transmission Decrease confirmed Evidence of no change Evidence of no change Evidence of no change Limited to clusters Similar to previous variants Data suggesting a difference Data suggesting a difference Data suggesting a difference Similar to previous variants Increase confirmed but limited to risk groups Experimental evidence of evasion Experimental evidence of evasion Experimental evidence of decreased efficacy Increase confirmed Increase confirmed for most groups Evidence of frequent reinfection Evidence of decreased efficacy Evidence of decreased efficacy
Genome maps
We (the Stanford HIVDB team) created these figures for different SARS-CoV-2 variants since this year, and we just released it under CC-BY-SA 4.0. I have uploaded the top four WHO named variants to Commons. Does anyone else other than me also think those figures are useful to be included in the variant articles? I'm afraid it is kind of interest conflict if I add those figures to the articles.
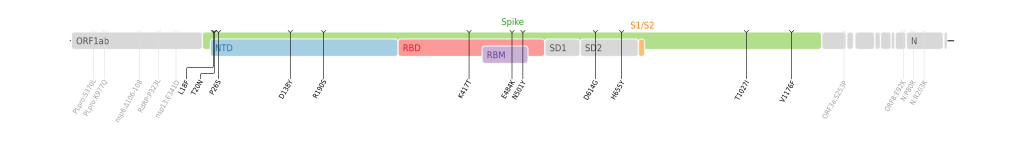
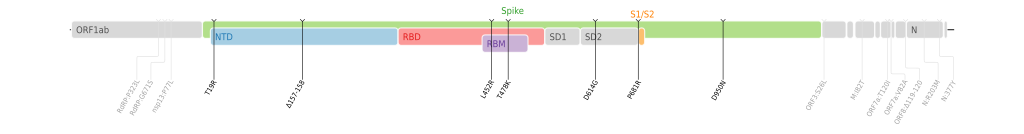
Best. --PhiLiP (talk) 19:06, 21 June 2021 (UTC)
- @PhiLiP: They seem pretty neat and useful. —hueman1 (talk • contributions) 08:36, 26 June 2021 (UTC)
- @HueMan1: Thank you very much! I have added more. Can you also help me to add them to the corresponding articles for me to avoiding conflicts of interest? Thanks!
- Best. --PhiLiP (talk) 17:35, 30 June 2021 (UTC)
- @PhiLiP: I've added the Kappa variant genome map to SARS-CoV-2 Kappa variant under the "mutations" section. These much-needed maps are excellent and so clear. Thank you for making them available. SpookiePuppy (talk) 01:08, 3 July 2021 (UTC)
- @PhiLiP: Genome maps now added to the remaining 6: SARS-CoV-2 Epsilon variant, SARS-CoV-2 Zeta variant, SARS-CoV-2 Eta variant, SARS-CoV-2 Theta variant, SARS-CoV-2 Iota variant and SARS-CoV-2 Lambda variant. SpookiePuppy (talk) 17:26, 5 July 2021 (UTC)
- @SpookiePuppy: Thank you very much! --PhiLiP (talk) 19:12, 5 July 2021 (UTC)
- @PhiLiP: Genome maps now added to the remaining 6: SARS-CoV-2 Epsilon variant, SARS-CoV-2 Zeta variant, SARS-CoV-2 Eta variant, SARS-CoV-2 Theta variant, SARS-CoV-2 Iota variant and SARS-CoV-2 Lambda variant. SpookiePuppy (talk) 17:26, 5 July 2021 (UTC)
- @PhiLiP: I've added the Kappa variant genome map to SARS-CoV-2 Kappa variant under the "mutations" section. These much-needed maps are excellent and so clear. Thank you for making them available. SpookiePuppy (talk) 01:08, 3 July 2021 (UTC)
grammar
"Limited evidence from various preliminary studies reviewed by the WHO have indicated" - is singular.
- I agree, if we take the object for this part of the sentence to be the word "evidence" (of "limited evidence"), then "have" needs to be changed to "has". Although, I should state that English grammar is not my strongest area. To simplify what's going on in that line, the first part effectively operates as: "Limited evidence...has indicated...". I'll go ahead and make the minor change, but if anyone disagrees, then feel free to just revert it. SpookiePuppy (talk) 20:07, 9 July 2021 (UTC)
- Follow-up: Having just tried to make the edit, it appears that the section of text cannot be edited from this page (Variants of SARS-CoV-2) as the text is being 'called' as a block from another page using a special Template:Excerpt. The page where the text can be edited is: COVID-19 vaccine. I'll make the edit there and leave a link to this talk page in the edit summary. SpookiePuppy (talk) 20:30, 9 July 2021 (UTC)
"B.1.429" listed at Redirects for discussion
![]() A discussion is taking place to address the redirect B.1.429. The discussion will occur at Wikipedia:Redirects for discussion/Log/2021 July 19#B.1.429 until a consensus is reached, and readers of this page are welcome to contribute to the discussion. ~~~~
A discussion is taking place to address the redirect B.1.429. The discussion will occur at Wikipedia:Redirects for discussion/Log/2021 July 19#B.1.429 until a consensus is reached, and readers of this page are welcome to contribute to the discussion. ~~~~
User:1234qwer1234qwer4 (talk) 11:24, 19 July 2021 (UTC)
Variant naming
I've been hearing about the "Delta variant" in the news recently and wanted to learn more about variants so I checked here, because Wikipedia generally does a good job of amalgamating information into one spot, especially for current events. I came to this page and became confused because the variants are not identified by their common name such as UK variant, South African variant, etc. Reading further, I can surmise which is possibly which BUT that is how I and many others know their names because that what the news calls/called them. At the very least there should be an also known as in the first sentence but there is not. I read some move requests and see this is largely due to not promoting a stigma. Wikipedia does a disservice to readers by referring to some variants solely by some language that is esoteric to most readers. This is a tertiary source and should cover things plainly. I don't edit on WP as much as I used to and from what I understand/see, it's fallen further off the deep end but I just wanted to state my opinion on how silly this is.--NortyNort (Holla) 12:55, 11 July 2021 (UTC)
- Since the WHO introduced the new names,[2] pretty much everyone has adopted them, from reporters to researchers. The only name deemed inadequate in Wikipedia is the stigmatizing one based on nationality/origin. This article actually goes to great lengths to associate a variant with the location of its first outbreak, to establish this connection in an ethical way. Before that change, the main name in Wikipedia was the Pango lineage name, which is far less friendly. For example, at the moment, everyone is reporting about the Delta variant, not about the Indian variant. Likewise, since the change happened recently, very little news has referred to the Lambda variant as the Andean variant. Labeling variants by origin quickly becomes impracticable because more than one variant can emerge in the same location, as is the case of Kappa in India. --Fernando Trebien (talk) 17:32, 11 July 2021 (UTC)
- I had a feeling they didn't put countries of names in each variants even if it's from Wikipedia. I know there be worries of stigmatizing on nationality or origins but we all pretty much know they all have it's roots and origin from the first Covid 19 variant which was the China/Wuhan typ. So people should know all these Indian variant, Peru variant, or Kappa variants, South Africa variants, American variants, UK variants, Belgium variants, Beta variants are all essentially a evolution from the first virus.. Although I can't complain about wikipedia not naming the countries origin of the virus. A lot of Indians had reported racism and worries because they called it the Indian variant(aka Delta variant) and I do admit this made people wanted to distance themselves from Indians. Vamlos (talk) 09:16, 17 July 2021 (UTC)
- Kappa is also endemic in India, so you end up with two "Indian" variants, and new variants are popping up all the time across the planet. Looking to the future, it's more about pragmatics than ethics. The new naming scheme actually appears to be based on the one used for types of influenzaviruses. --Fernando Trebien (talk) 20:52, 22 July 2021 (UTC)
- I had a feeling they didn't put countries of names in each variants even if it's from Wikipedia. I know there be worries of stigmatizing on nationality or origins but we all pretty much know they all have it's roots and origin from the first Covid 19 variant which was the China/Wuhan typ. So people should know all these Indian variant, Peru variant, or Kappa variants, South Africa variants, American variants, UK variants, Belgium variants, Beta variants are all essentially a evolution from the first virus.. Although I can't complain about wikipedia not naming the countries origin of the virus. A lot of Indians had reported racism and worries because they called it the Indian variant(aka Delta variant) and I do admit this made people wanted to distance themselves from Indians. Vamlos (talk) 09:16, 17 July 2021 (UTC)
References
- ^ "SARS-CoV-2 variants: risk assessment framework" (PDF). GOV.UK. Government Digital Service. Public Health England. 2021-05-22. GOV-8426.
- ^ "Covid: WHO renames UK and other variants with Greek letters". BBC. 31 May 2021.
"relative to Alpha" fixings
I'm well aware that most cited sources will place the benchmark of Delta's stats relative to Alpha, but if only for the sake of curiosity I am wondering how we can extrapolate its changes relative to the wild strains from this data. Apparently simply doing the maths on the data (ex. since Delta is 64% more transmissible than Alpha, and Alpha itself is 82% more transmssible, it would seem to calculate Delta's transmissibility compared to the wild strains you'd just do 1.64 * 1.82 = 2.98 = +198%, and do the same to the range calculations based on Delta's hypothesized error margin) does not actually yield the correct percentage, as many here have pointed out, but aside from that I'm totally ignorant on what the correct course of action is. It may not seem like a huge issue to leave Delta's stats as relative to Alpha, and I'm not trying to suggest this even is an issue to begin with, but it's my personal opinion that we 'ought to keep consistency and make all variants in the overview table show their changes relative to wild COVID-19. 47.20.177.163 (talk) 16:44, 22 July 2021 (UTC)
- We must report the best information available. To properly calculate confidence intervals, it is necessary to have access to research data. Verifying the correctness of this calculation is an important step in peer review, so doing this on Wikipedia would be similar to original research, which does not belong to Wikipedia. Reporting a relative change is not really a huge issue and will likely happen again with new variants of concern. When a variant emerges, researchers are initially able to measure only relative behavior in the region of the outbreak, and establishing a comparison to the original variant requires complicated modeling and is not really useful in a vaccinated population (it is useful in an unvaccinated population). You see, for example, that most research on Alpha comes from the UK, which was hit first. Likewise, there is almost no research on Beta and Gamma, which are rare in wealthy countries able to sponsor research, so naturally everyone is prioritizing their most immediate risks, while this table tries to represent global risks that could hit everyone, some time in the future. I can also mention the case of Epsilon, which was downgraded from VOC to VOI by the CDC because it was overtaken by more transmissible variants, so its risk was evaluated in a relative perspective as well. This preprint is already used to fill one of the cells that was empty until recently, and it has data for hospitalization, so when it is published it may replace another cell. Then there is transmissibility, for which there is no other reliable source than those already cited. --Fernando Trebien (talk) 20:13, 22 July 2021 (UTC)
- Fair enough. I realized the problems with deviating from the cited sources, even if we were simply restating their information in a different way, midway through writing this anyway. 47.20.177.163 (talk) 17:18, 23 July 2021 (UTC)
Mechanism for infection peaks
This article doesn't really provide any mechanism to explain how variants remain statistically dormant for many months, and then allegedly correlate with massive unprecedented infection peaks in the nations where they are first identified, more or less immediately after the vaccination process begins. Since this is a pervasive pattern among these variants, it demands at least some kind of explanation. - Joshua Clement Broyles
- A variant simply emerges from one or more mutations. It can infect its host or it can infect other people it comes in contact with. From there, it spreads following its own transmission rate, which is not known until studies are done on a large scale (hence the unpredictability). You simply have to do the math of exponential functions to see how long it takes for a case to become millions and surpass the ones that already occur. Things like seasonality (going through summer), interventions (lockdowns, social distancing, wearing masks) and cross-reactivity (immunity to infections by other variants) can slow this process at varying rates over time, increasing unpredictability. --Fernando Trebien (talk) 17:24, 23 July 2021 (UTC)
COVID-19 Variants Update
By the New Zealand Ministry of Health
- https://www.health.govt.nz/our-work/diseases-and-conditions/covid-19-novel-coronavirus/covid-19-resources-and-tools/covid-19-science-news#variants
- 11 August 2021 - Variants Update - Full Report (PDF, 1.02 MB)
- 18 August 2021 - Variants Update - Full Report] (PDF, 663 KB)
Yug (talk) 16:59, 28 August 2021 (UTC)
Circular links
I removed a bunch of circular links in the section https://en.wikipedia.org/wiki/Variants_of_SARS-CoV-2#Additional_variants . Per MOS:CIRCULAR circular links are a no no. Using common sense, circular link are not helpful. Please do not put circular links in the future. Daniel.Cardenas (talk) 03:50, 1 September 2021 (UTC)
B.1.621
WHO has added "Mu" (Pango lineage B.1.621) as a Variant of Interest. Would anyone like to add this to the wiki article?
https://www.ecdc.europa.eu/en/covid-19/variants-concern
https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ Hongsy (talk) 04:21, 1 September 2021 (UTC)
P1 > Assessment source
- Freitas, André Ricardo Ribas; Lemos, Daniele Rocha Queiróz; Beckedorff, Otto Albuquerque; Cavalcanti, Luciano Pamplona de Góes; Siqueira, Andre M.; Mello, Regiane Cristina Santos de; Barros, Eliana N. C. (2021-04-19). "The increase in the risk of severity and fatality rate of covid-19 in southern Brazil after the emergence of the Variant of Concern (VOC) SARS-CoV-2 P.1 was greater among young adults without pre-existing risk conditions". medRxiv: 2021.04.13.21255281. doi:10.1101/2021.04.13.21255281.
Delta mortality: CFR in England not applicable
Overview table
It would be helpful if a row for the original virus could be added to the top of the table. I don't have the medical qualifications to do that. Lay people may assume the Alpha variant is the original virus unless they read the article carefully. Ghostofnemo (talk) 02:25, 22 July 2021 (UTC)
- The table describes changes from the original variant and those similar to it. Adding a line for the original variant only makes sense if we describe absolutes rather than relatives, such as reproduction number (R0 or Rt) rather than changes in transmissibility. So far, most of the data are being reported relative to the original variant because it is the one most governments have prepared for. Of course that will likely change. In the UK, for example, most reports now relate to Alpha because it has surpassed all other strains that behaved like the original. Soon, unfortunately, the reporting will likely establish Delta as the main reference. --Fernando Trebien (talk) 20:42, 22 July 2021 (UTC)
- @Ftrebien: Would you consider a supplementary table include the absolute values to be a violation of the policy against original research? I'm not sure about that, which is why I'm hesitant to add a supplementary table. CessnaMan1989 (talk) 01:43, 5 November 2021 (UTC)
- @CessnaMan1989: After this question was raised, I found and added a better reference with values relative to the original variant, so I don't think this is a problem anymore. But suppose we were still back on 22 July. The problem is that we would be composing two somewhat uncertain measurements to arrive at a new and less certain measurement. We should ask ourselves why researchers are publishing values relative to variants currently in circulation, rather than absolute values that would be comparable to the original variant. The researchers from this new source I found had direct access to the data to recalculate the confidence intervals, we in Wikipedia do not have that access and if we tried to do the same, it would be the same as taking the role of those researchers. So, yes, I think this would qualify as original research. The best we could do is multiply the two estimates, but then we would be unable to provide a confidence interval, so the reader wouldn't know how accurate the composite estimate is. That might be acceptable, but it would be better to add a note explaining where the composite value came from. --Fernando Trebien (talk) 12:26, 5 November 2021 (UTC)
Medrxiv sourcing and technical DOI issues
@Jaredroach: I fully understand medrxiv is not a proper Wikipedia reference. I'm glad you've decided to remove medrxiv references which are not based on any peer review. However, one of the articles you removed is actually also published in a reputable journal (Cell (journal)), and should have been updated instead of removed.
The point of my edit was to add in a DOI pointing to the peer reviewed paper. Before my edit, the article had a DOI pointing to the medrxiv preprint, but not the actual paper. Unfortunately, the Template:cite journal does not have a parameter for medrxiv preprints within the same citation, so I moved the preprint link to a separate template within the same footnote to keep the source accessible, since the publisher article is paywalled. I'm not adding any references here, just clarifying the DOI of the authoritative source.
I think your revert was based on a misunderstanding. Finding an article on medRxiv doesn't mean it's *not* peer reviewed, it's just not a sufficient evidence of peer review. 〈 Forbes72 | Talk 〉 22:10, 2 October 2021 (UTC)
- That's fine. But in no case should there be any need to reference medrxiv. If an article in medrxiv is published later in a reputable source, one should reference the reputable source. One should not reference medrxiv "pending" the availability of that reputable source. I think it is correct for one editor to remove the medrxiv source, and for another editor to add the new source. The first editor might not easily have the info for next source, nor should they feel obligated to find it before deleting the medrxiv source. After removing the medrxiv source, the material may be unsourced. A "citation needed" tag can be added, and eventually the material can be deleted or someone can add the new reference. Jaredroach (talk) 22:16, 2 October 2021 (UTC)
- I agree with you that using preprints as "pending" sources should never be done, and any non-peer reviewed sources used as a reference should be deleted. I'd just point out that arxiv/biorxiv/medrxiv will link from the preprint directly to the published version if it exists, so it's a one-click check to see if the reference is salvageable. I assume you're just objecting to standalone medrxiv references as bad, and not any and all preprint links? Adding a link to the preprint is accepted practice on Wikipedia as long as the underlying peer-reviewed paper is also linked in the same citation. I three options to add the missing peer-reviewed link to the citation:
- 1. replace the DOI link to the preprint with a DOI link to the reviewed paper, but keep the preprint accessible in a nearby template (what I did before revert)
- 2. replace the DOI link to the preprint with a DOI link to the reviewed paper, remove the link to the preprint entirely (I would object if medrxiv was the only free source, but since there's a PMC link this doesn't matter much)
- 3. update the cite journal template so we can include both the DOI link to the paper and DOI link to the preprint in the same template.
- I'd say option 3 is ideal, but it would take some time since cite journal is a highly visible, fully protected template. What do you think? 〈 Forbes72 | Talk 〉 23:26, 2 October 2021 (UTC)
- Absent explicit assurance that the content of the preprint is identical with the content of the peer-reviewed (published) version, they should not be considered equivalent. Peer review often results in revisions (it's not simply an approval process). — soupvector (talk) 23:42, 2 October 2021 (UTC)
- That's a very technical distinction to make. In my experience comparing peer-reviewed papers to preprints, preprints verus published paper end up pretty similar, so I usually prefer to link to both in the case of a paywall. I agree the published one is the authoritative source. I think its a mistake to remove preprint links for peer-reviewed papers, but I've gone ahead and removed the preprint doi altogether in this one case so at least the citation template is correct. 〈 Forbes72 | Talk 〉 01:59, 31 October 2021 (UTC)
- No idea how easy it is to update MedrXiv preprints, but updating ArXiv preprints to match the final accepted version (in content, not format, font, etc) is standard practice. This guarantees green open access. Any non-trivial changes in the review process are normally put in the final versions of the preprints, because it's a lot easier to check ArXiv preprints than waste time trying to guess which journals are non-paywalled. Moreover, some papers have mathematical errors introduced by proofreaders that the authors fail to pick up when double-checking the proofs, in which case the version-of-record is wrong and the final preprint is right. The peer review process statistically improves the quality of research papers, but hiding or removing preprint information generally functions as a block against access to verifiable information, especially with journals that are still paywalled. Caveat: the actual practice may vary across disciplines - here I'm talking of astronomy/cosmology - I can't judge MedrXiv. Boud (talk) 06:51, 26 November 2021 (UTC)
- That's a very technical distinction to make. In my experience comparing peer-reviewed papers to preprints, preprints verus published paper end up pretty similar, so I usually prefer to link to both in the case of a paywall. I agree the published one is the authoritative source. I think its a mistake to remove preprint links for peer-reviewed papers, but I've gone ahead and removed the preprint doi altogether in this one case so at least the citation template is correct. 〈 Forbes72 | Talk 〉 01:59, 31 October 2021 (UTC)
- Absent explicit assurance that the content of the preprint is identical with the content of the peer-reviewed (published) version, they should not be considered equivalent. Peer review often results in revisions (it's not simply an approval process). — soupvector (talk) 23:42, 2 October 2021 (UTC)
- I agree with you that using preprints as "pending" sources should never be done, and any non-peer reviewed sources used as a reference should be deleted. I'd just point out that arxiv/biorxiv/medrxiv will link from the preprint directly to the published version if it exists, so it's a one-click check to see if the reference is salvageable. I assume you're just objecting to standalone medrxiv references as bad, and not any and all preprint links? Adding a link to the preprint is accepted practice on Wikipedia as long as the underlying peer-reviewed paper is also linked in the same citation. I three options to add the missing peer-reviewed link to the citation:
Proposal to add mutations to variant tables (total and in the spike protein)
Especially regarding the news for Variant B.1.1.529 with 32 mutations in the spike protein (for comparison Delta has 7+) also found in an Israeli Tourist returning from Malawi probably boostered with Pfizer–BioNTech[1] it should be informative to add the info of known total and spike protein mutations to all variant comparison tables, so under Overview variants of concern (VOC) and Alerts for further monitoring (WHO) and maybe if possible also under Nomenclature SARS-CoV-2 corresponding nomenclatures. Understanding the range of mutational variability while remaining infectious of the SARS-CoV-2 virus is of utter importance combined with understanding how it effects natural and vaccine immunity in the variants of concern. --Krahax (talk) 08:54, 26 November 2021 (UTC)
- The VOC table is a bit overloaded, but the Alerts table has plenty of space. However, being so atypical for B.1.1.529, I think it would be best to write this in prose for due weight. The number of mutations has theoretical implications - it "may" affect immunity from vaccines and the effectiveness of antibody therapies; but it also may not. Most mutations do not affect these characteristics, some do significantly, and the combined effect is difficult to predict. --Fernando Trebien (talk) 12:18, 26 November 2021 (UTC)
- Thx for the response Ftrebien. AFAIK the immunity could be much more effected by the number of mutations in contrast to transmission-rate and severity which should depend more on the type of mutations. But of course any paper/expert review on this is welcome. I can't find the source but found an interesting 2D/3D plot of the mutation differences of Alpha, Beta, Gamma, Delta interesting. I'm not sure if it was in a paper with CC-BY(-SA) licence or in a media article. Not sure if possible but a comparison of the predicted post-Delta Scenarios would be informative to see if one model is close to the future reality. --Krahax (talk) 13:06, 26 November 2021 (UTC)
"Nu" Omicron variant
There are lots of mainstream news reports stating that B.1.1.529 has been named as the "nu" variant by the WHO. However, this seems to be jumping the gun; I see that the meeting to discuss this is happening right now (see [7]), and there is as yet no official word on this. As or when a "nu" variant is declared in WP:MEDRS, it should be added here. -- The Anome (talk) 17:03, 26 November 2021 (UTC)
- And here we go: WHO has indeed declared B.1.1.529 a variant of interest, (see [8]) but assigned it the Greek letter omicron, not nu. Perhaps because "nu" sounds like "new"? -- The Anome (talk) 18:11, 26 November 2021 (UTC)
- If they are skipping nu, the next should be xi not omicron. Agmartin (talk) 18:15, 26 November 2021 (UTC)
- WHO published a statement about classifying B.1.1.529 as Omicron and a Variant of Concern.[2] ― Aidan ⦿ (talk) 18:33, 26 November 2021 (UTC)
WHO recently named the new variant lineage #B11529 as "Omicron," but I have not been able to find here any info on the variant named "Xi", which is the preceding letter ib the Greek alphabet. 108.12.194.196 (talk) 19:00, 26 November 2021 (UTC)
- Given that China's current leader is Xi Jinping, I think it's quite unlikely that the WHO will use the letter. I'm also guessing that it's likely that they skipped "nu" because it sounded too much like "new". -- The Anome (talk) 19:04, 26 November 2021 (UTC)
References
- ^ "Statement from Israel's health ministry reporting 1 confirmed case of new coronavirus variant B.1.1.529 [unofficial translation]". @BNODesk Twitter. Retrieved 26 November 2021.
- ^ "Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern". WHO.int. World Health Organization. Archived from the original on 26 Nov 2021. Retrieved 26 Nov 2021.
WHO skips nu and xi
WHO skips nu and xi.
- https://www.rt.com/news/541479-who-nu-xi-omicron-covid/
- https://nypost.com/2021/11/26/who-skips-two-letters-in-alphabet-in-naming-omicron-variant/ — Preceding unsigned comment added by 208.98.202.34 (talk) 05:46, 27 November 2021 (UTC)
- yes your correct, rather odd on their part--Ozzie10aaaa (talk) 13:16, 28 November 2021 (UTC)
B.1.1.529
Just saw news of this breaking over the last couple of hours. It's currently listed on the WHO tracking page as a variant under monitoring. I'm not sure what counts as a RS in this topic area yet, I note however that Axios have stated that it's ben found so far in Botswana, South Africa, and Hong Kong. Reuters state 100 specimens in South Africa, and confirm what Axios state for Botswana and Hong Kong but state the Hong Kong case is a traveller from South Africa. Sideswipe9th (talk) 18:19, 25 November 2021 (UTC)
- I've added the basic info from the WHO tracking page. Not sure what to cite for more detailed information in the notes section however. Sideswipe9th (talk) 18:25, 25 November 2021 (UTC)
- Thanks User:Sideswipe9th for making the addition. I think the sheer number of mutations on the spike protein alone is the most concerning part and perhaps needs to be mentioned.[1] The following news sources state that there are 32 spike mutations.[2][3] SpookiePuppy (talk) 19:45, 25 November 2021 (UTC)
- @PhiLiP: I've just had an afterthought, to ping the Stanford HIVDB team who created those superb genome map figures (https://en.wikipedia.org/wiki/Talk:Variants_of_SARS-CoV-2#Genome_maps) which showed the different SARS-CoV-2 variants, to make them aware of this discussion about the new variant (B.1.1.529). SpookiePuppy (talk) 20:14, 25 November 2021 (UTC)
- @SpookiePuppy: Thanks! We are updating our site correspondingly. --PhiLiP (talk) 18:10, 28 November 2021 (UTC)
- @PhiLiP: I've just had an afterthought, to ping the Stanford HIVDB team who created those superb genome map figures (https://en.wikipedia.org/wiki/Talk:Variants_of_SARS-CoV-2#Genome_maps) which showed the different SARS-CoV-2 variants, to make them aware of this discussion about the new variant (B.1.1.529). SpookiePuppy (talk) 20:14, 25 November 2021 (UTC)
- Thanks User:Sideswipe9th for making the addition. I think the sheer number of mutations on the spike protein alone is the most concerning part and perhaps needs to be mentioned.[1] The following news sources state that there are 32 spike mutations.[2][3] SpookiePuppy (talk) 19:45, 25 November 2021 (UTC)
- This reference[4] might be useful, but the situation at the moment is so uncertain that I think it would be better to wait for a statement from the WHO. The effect of these mutations on neutralization needs to be tested. --Fernando Trebien (talk) 00:26, 26 November 2021 (UTC)
- Six Southern African countries added to England's red list for travel; "100 cases confirmed through full sequencing of samples, up from just 10 reported on Wednesday, and signs of community transmission," 90% of cases in Gauteng.[5] But still only VUM for WHO, not (yet) VOI or VOC. Boud (talk) 01:01, 26 November 2021 (UTC)
- WHO will meet 26 Nov 2021 to decide if upgrading B.1.1.259 from VUM to VOI or VOC is justified.[6]
- Six Southern African countries added to England's red list for travel; "100 cases confirmed through full sequencing of samples, up from just 10 reported on Wednesday, and signs of community transmission," 90% of cases in Gauteng.[5] But still only VUM for WHO, not (yet) VOI or VOC. Boud (talk) 01:01, 26 November 2021 (UTC)
- There are already draconian travel restrictions all over the lot. Once WHO settles on the nomenclature, article drops. This thing came out of the gate fast...! https://www.theguardian.com/world/live/2021/nov/26/covid-news-live-new-variant-sparks-tougher-restrictions-in-india-and-singapore-ahead-of-who-meeting
kencf0618 (talk) 10:19, 26 November 2021 (UTC)
- David Nabarro from the WHO: "it is going to be some weeks before we can say for certain whether our fears have any basis" [9] --Fernando Trebien (talk) 14:44, 26 November 2021 (UTC)
- Maybe Responses to the COVID-19 pandemic in November 2021 is a good place to write about this subject for now. --Fernando Trebien (talk) 14:53, 26 November 2021 (UTC)
- Certainly the UK's addition to their travel red list should be added to that page. Sideswipe9th (talk) 15:50, 26 November 2021 (UTC)
- I've added a quick and dirty prècis focusing on Africa, but note that the impact is already global. It'll get its own article and timeline soon enough. (I refer to it as "Variant Nu" inasmuch as WHO hasn't officially announced the nomenclature.) kencf0618 (talk) 17:24, 26 November 2021 (UTC)
Hong Kong
This edit request has been answered. Set the |answered= or |ans= parameter to no to reactivate your request. |
In the article Hong Kong is said to have reported one case. There have actually been two, from the very moment it was announced that the B.1.1.529 strain was detected in their samples.[10][11][12][13] 1.64.47.144 (talk) 14:37, 27 November 2021 (UTC)
 Not done. 'The Straits Times' reports that it was "brought in by a man who had flown in from South Africa on Nov 11... This patient, 36, then allegedly passed on the virus to another man who was staying in a neighbouring room at the hotel." I think it makes more sense to keep the article saying "one" since there was only one person that brought it to Hong Kong, and saying two would imply otherwise. ◢ Ganbaruby! (talk) 00:02, 3 December 2021 (UTC)
Not done. 'The Straits Times' reports that it was "brought in by a man who had flown in from South Africa on Nov 11... This patient, 36, then allegedly passed on the virus to another man who was staying in a neighbouring room at the hotel." I think it makes more sense to keep the article saying "one" since there was only one person that brought it to Hong Kong, and saying two would imply otherwise. ◢ Ganbaruby! (talk) 00:02, 3 December 2021 (UTC)
Xi variant is not a "common" last name
It should be noted in the article that Xi is factually not a very "common" last name. While the threshold of "common" can be debated, the point is entirely moot as stated. The surname Mu, an already named variant recognized by WHO, is relatively much more common of a last name.
MU: ~1025k instances, 527th most common (https://forebears.io/surnames/mu)
XI: ~775k instances, 708th most common (https://forebears.io/surnames/xi)
The fact that the current CCP leader's name is Xi, however, remains factually correct. — Preceding unsigned comment added by 73.8.240.69 (talk) 03:32, 4 December 2021 (UTC)
- According to China's Ministry of Public Security http://gat.ah.gov.cn/public/7081/40201012.html via Google Translate there are "more than 6,000 surnames in use today" (though https://edition.cnn.com/2021/01/16/china/chinese-names-few-intl-hnk-dst/index.html, based on that has "about 6,000 surnames are in use". According to https://www.scmp.com/news/china/science/article/3157678/lambda-mu-omicron-why-who-skipped-nu-and-xi-name-latest, based on the Ministry of Public Security data, "the Chinese president’s surname is the 296th most common family name" and "Two other surnames that would also be read as “Xi” but read in different tones were more common, ranking 169 and 228". Mcljlm (talk) 08:01, 4 December 2021 (UTC)
"Positive" vs. "Negative" vs. "Neutral" mutations
Could somebody further explain to me what the video in the article means by "positive mutations" and "negative mutations"? What makes a mutation positive or negative? CessnaMan1989 (talk) 01:48, 5 November 2021 (UTC)
- Google scholar might help--Ozzie10aaaa (talk) 15:23, 4 December 2021 (UTC)
Original strain name
Is there a name for the original strain, eg. "SARS‑CoV‑2 wild-type" or "SARS‑CoV‑2 original" or something like that? Or is its name just "SARS‑CoV‑2", without any qualifiers? There are some older designators, like SARS‑CoV‑2 strain G and SARS‑CoV‑2 strain L; are these differences regarded as being on the same level as the Greek-lettered variants, or as just minor differences within a single variant? -- The Anome (talk) 13:16, 3 December 2021 (UTC)
- I could be wrong but I believe its SARS‑CoV‑2--Ozzie10aaaa (talk) 15:25, 4 December 2021 (UTC)
Nomenclature
Currently the 2nd sentence of Section #2 Nomenclature is "As of July 2021, no consistent nomenclature was established for it." whose reference (via Twitter) is a January 2021 WHO statement archived from the original on 23 January 2021, retrieved 2 February 2021. Since a later sentence is "After months of discussions, the World Health Organization announced Greek-letter names for important strains on 31 May 2021." (also with a Twitter reference) should "July" be a different month? Maybe https://www.who.int/publications/i/item/WHO-2019-nCoV-genomic_sequencing-2021.1 should be used for the January reference and https://www.who.int/news/item/31-05-2021-who-announces-simple-easy-to-say-labels-for-sars-cov-2-variants-of-interest-and-concern for the May reference instead of Twitter. Mcljlm (talk) 00:56, 1 December 2021 (UTC)
- thank you for post--Ozzie10aaaa (talk) 15:26, 4 December 2021 (UTC)
Source for risk classification
The table under Overview highlights many cells according to some color coding for risk. Is there any source for that, or is it OR? ◅ Sebastian 12:37, 29 November 2021 (UTC)
- how would 'color coding' be OR?--Ozzie10aaaa (talk) 15:25, 4 December 2021 (UTC)
- @Ozzie10aaaa:: Simple: When someone marks a variant with a color such as red, it means “Very high risk”. That's a statement of fact – and even an extreme and potentially dangerous one – that obviously needs a reference. ◅ Sebastian 22:22, 3 January 2022 (UTC)
New variant detected in France (B.1.640.2)
There are news reports of another variant being detected in Forcalquier in the south of France.[7][8] It has been provisionally named B.1.640.2. Apparently it was detected in early December 2021 at the IHU Méditerranée, Forcalquier. Scientists are suggesting that the origin may lead back to a person returning from a trip to the Cameroon.[9] SpookiePuppy (talk) 21:03, 3 January 2022 (UTC)
- And it's likely to become "variant of concern"[10] -Abhishikt (talk) 00:57, 4 January 2022 (UTC)
- I've created a draft at Draft:SARS-CoV-2 lineage B.1.640.2; please feel free to expand. Tol (talk | contribs) @ 01:49, 5 January 2022 (UTC)
A new variant B.1.640.2 aka "IHU" has emerged in France that's more infectious than SARS-CoV-2 Omicron variant. Please find the details in the references.[11][12][13] — Preceding unsigned comment added by 2402:3a80:6f2:f627:7904:c8b8:6239:7bd1 (talk • contribs)
- Comment. Someone has created an article named B.1.640.2. Maybe it must be deleted/redirected. Alexcalamaro (talk) 12:33, 5 January 2022 (UTC)
- There's also the draft Draft:SARS-CoV-2 lineage B.1.640.2 mentioned above, which is a little more developed than B.1.640.2. Perhaps B.1.640.2 should be nominated for deletion? It's also a bit early for this variant to have its own page, although the variant is not as new as it might seem. SpookiePuppy (talk) 14:47, 5 January 2022 (UTC)
- The article has already been edited by Valepert (talk · contribs) to a redirect to this article. So I have added a "R with possibilities" template, and created a draft article with the name "Draft:B.1.640.2" with a redirect to "Draft:SARS-CoV-2 lineage B.1.640.2". Alexcalamaro (talk) 18:02, 5 January 2022 (UTC)
- There's also the draft Draft:SARS-CoV-2 lineage B.1.640.2 mentioned above, which is a little more developed than B.1.640.2. Perhaps B.1.640.2 should be nominated for deletion? It's also a bit early for this variant to have its own page, although the variant is not as new as it might seem. SpookiePuppy (talk) 14:47, 5 January 2022 (UTC)
- A variant found in France is not a concern, the W.H.O. says.[14] -Abhishikt (talk) 23:40, 5 January 2022 (UTC)
References
- ^ https://github.com/cov-lineages/pango-designation/issues/343
- ^ "Great news! A new horrifying SARS-CoV-2 variant B.1.1529 with a high number of worrying mutations has emerged in time for the festive season!". Thailand Medical News. Thailand. 25 November 2021. Retrieved 2021-11-25.
- ^ Sample, Ian (24 November 2021). "Scientists warn of new Covid variant with high number of mutations". The Guardian. London. Retrieved 2021-11-25.
- ^ Callaway, Ewen (2021-11-25). "Heavily mutated coronavirus variant puts scientists on alert". Nature. doi:10.1038/d41586-021-03552-w.
- ^ https://www.theguardian.com/world/2021/nov/25/scientists-call-for-travel-code-red-over-covid-variant-found-in-southern-africa
- ^ https://www.theguardian.com/world/2021/nov/26/who-to-assess-new-highly-mutated-covid-19-variant-as-countries-ramp-up-health-checks
- ^ https://www.lematin.ch/story/nouveau-variant-decouvert-dans-le-sud-de-la-france-626037426073
- ^ https://www.jpost.com/health-and-wellness/coronavirus/article-691458
- ^ https://www.medrxiv.org/content/10.1101/2021.12.24.21268174v1
- ^ https://www.examinerlive.co.uk/news/uk-world-news/new-variant-france-46-mutations-22634183
- ^ Chaturvedi, Amit (4 January 2022). "New Covid-19 variant 'IHU' discovered in France, has more mutations than Omicron". Hindustan Times.
- ^ "COVID-19: New variant, B.1.640.2, detected in France - study". The Jerusalem Post. Retrieved 4 January 2022.
- ^ Colson, Philippe; Delerce, Jérémy; Burel, Emilie; Dahan, Jordan; Jouffret, Agnès; Fenollar, Florence; Yahi, Nouara; Fantini, Jacques; La Scola, Bernard; Raoult, Didier (29 December 2021). "Emergence in Southern France of a new SARS-CoV-2 variant of probably Cameroonian origin harbouring both substitutions N501Y and E484K in the spike protein". medRxiv. doi:10.1101/2021.12.24.21268174.
- ^ https://www.nytimes.com/2022/01/05/world/covid-variant-france.html
Merge of B.1.640.2
The following discussion is closed. Please do not modify it. Subsequent comments should be made on the appropriate discussion page. No further edits should be made to this discussion.
I've proposed a merge of SARS-CoV-2 lineage B.1.640.2 here. Also see Draft:SARS-CoV-2 Ihu variant. B.1.640.2 is not a variant of concern, nor even a variant of interest. The sourcing is not really sufficient, based as it is in a preprint. See also Wikipedia_talk:New_pages_patrol/Reviewers#MEDRS_issue. Fences&Windows 10:41, 6 January 2022 (UTC)
- Support this merge. (t · c) buidhe 10:54, 6 January 2022 (UTC)
- Support to merge (if its status changes in the future we'll see, but right now lets merge them). Alexcalamaro (talk) 12:07, 6 January 2022 (UTC)
- support per Alexcalamaro--Ozzie10aaaa (talk) 15:04, 6 January 2022 (UTC)
- Sure, I don't mind. Tol (talk | contribs) @ 19:17, 6 January 2022 (UTC)
- Support the proposal to merge B.1.640.2 into Variants of SARS-CoV-2 as there are several reasons why it should not have its own page right now, especially the limited references. SpookiePuppy (talk) 14:42, 7 January 2022 (UTC)
- Support as per SookiePuppy HurricaneEdgar 12:00, 9 January 2022 (UTC)
- Support -- Aquaphoton (talk) 18:26, 11 January 2022 (UTC)
Thanks for the message on my talk page. I edited my draft for Draft:SARS-CoV-2 Ihu variant, as I realized that IHU was first detected in November. Also made other minor edits. Please help improve the references. Aquaphoton (talk) 18:16, 11 January 2022 (UTC)
D614 > G614 mutation in February 2020?
In this International Journal of Clinical Practice article ( https://pubmed.ncbi.nlm.nih.gov/32374903/ ) and in this Cell (journal) article ( https://pubmed.ncbi.nlm.nih.gov/32697968/ ) the authors describe a mutation of the original wuhan strain in position 614 where aspartate is replaced by glycine. This mutation likely happened in February in Italy and it apparently took over rapidly: "The shift occurred even in local epidemics where the original D614 form was well established prior to introduction of the G614 variant. The consistency of this pattern was highly statistically significant, suggesting that the G614 variant may have a fitness advantage." (Abstract of the Cell paper). Shouldn't this be mentioned somewhere in the article? I know this is long before the current "variants of concern" nomenclature, which began with the Alpha variant first observed in Kent in late 2020, but still, G614 is a variant, and it (according to the papers) seems to have behaved significantly different. Cheers --91.64.58.74 (talk) 19:28, 22 January 2022 (UTC)
- See Variants of SARS-CoV-2#D614G. The D614G clade, B.1, includes the major variants Alpha, Beta, and Omicron. Fences&Windows 00:21, 23 January 2022 (UTC)
Deltacron variant
Here is an article on the new Deltacron variant: [14] Yodabyte (talk) 03:39, 15 January 2022 (UTC)
- Promptly debunked as lab contamination: https://www.nature.com/articles/d41586-022-00149-9 Fences&Windows 01:26, 22 January 2022 (UTC)
- A genuine Deltacron genome has now been sequenced: [15] —Legoless (talk) 16:52, 11 March 2022 (UTC)
- https://www.theguardian.com/world/2022/mar/11/what-is-deltacron-covid-variant-uk also reports this as DIFFERENT from the earlier deltacron - yet this is not discernable from wikipedia as sometime it has been deleted so there is no information. 88.112.31.26 (talk) 03:28, 12 March 2022 (UTC)
- A genuine Deltacron genome has now been sequenced: [15] —Legoless (talk) 16:52, 11 March 2022 (UTC)
Variants under monitoring: XD
https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ now lists XD (Pango lineage https://cov-lineages.org/lineage.html?lineage=XD ) 92.18.224.107 (talk) 12:04, 29 March 2022 (UTC)
 Partly done
Partly done Done XD variant added.
Done XD variant added. Tables need to be updated for former monitored variants (I haven't been able do edit them from my mobile). Tables updaded Alexcalamaro (talk) 12:57, 12 April 2022 (UTC)
BA.4 and BA.5
https://www.forbes.com/sites/brucelee/2022/05/16/european-cdc-calls-omicron-ba4-ba5-covid-19-variants-of-concern/?sh=173c4db624a5 should they be mentioned here too? — Preceding unsigned comment added by 88.112.31.26 (talk) 03:33, 20 May 2022 (UTC)
 Done I think they deserve to be mentioned under the sublineages of Omicron, given that the WHO [16], the ECDC [17], and the CDC [18] all list BA.1 to BA.5 as variants of concern (with the exception of the ECDC for BA.3). EduardoW (talk) 20:42, 24 May 2022 (UTC)
Done I think they deserve to be mentioned under the sublineages of Omicron, given that the WHO [16], the ECDC [17], and the CDC [18] all list BA.1 to BA.5 as variants of concern (with the exception of the ECDC for BA.3). EduardoW (talk) 20:42, 24 May 2022 (UTC)
Add a Current Event template?
Given the nature of the frequently changing dominating variants, I would like to discuss the pros and cons of adding a Current events template. I admit that the template does not fit perfectly - after all, the list of variants is not an "event" in and of itself, but I think it's the closest we have to stating "This page's contents changes frequently and readers should verify themselves if the information are up to date". Thoughts on this? EduardoW (talk) 05:56, 26 May 2022 (UTC)
Cleanup discussion (updated 2022-07-11)
I personally think the article has gotten too long and should be cut down significantly, especially concerning variants that no longer circulate or never circulated significantly. However, before deleting large amounts of text, I would like to initiate a discussion here.
- Should we shorten the texts on the previously circulating VOCs and VOIs? In particular: Gamma, Delta, Epsilon
- Some suggestions for the large, colored table:
- We could make it collapsible (see Special:PermanentLink/1094560249). MovieboxFan, you did not like this, and I partially agree with your points.
- Pro: All the information is still there, the article gets a lot shorter
- Contra: Harder to find
- We could relocate the table to Previously circulating variants of concern (VOC), since 80 % of its information is on such VOCs
- Pro:
- Cleans up the start of the article
- More currently relevant information in the first parts of the article
- Contra:
- The Omicron information does not really make sense there
- We could make it collapsible (see Special:PermanentLink/1094560249). MovieboxFan, you did not like this, and I partially agree with your points.
I'd appreciate your opinions on this. EduardoW (talk) 06:20, 11 July 2022 (UTC)
- thank you for post--Ozzie10aaaa (talk) 17:52, 31 October 2022 (UTC)
XBB
The article mentions sub-variants BA.5 and BA.2.75; shouldn't it also mention XBB, which is quickly displacing BA.5 and BA.2.75 in places such as Singapore?[1] 2601:281:D47E:1920:5122:42BB:EED0:F063 (talk) 04:48, 17 October 2022 (UTC)
References
suggestion to use ISO 8601 date formats so dates will sort correctly
This page has several tables containing dates. However, the dates in the tables do not sort reliably due to the way they are currently formatted. Reformatting the dates using the ISO 8601 date format would resolve the date sorting issue. 76.92.203.206 (talk) 20:29, 14 November 2022 (UTC)
Semi-protected edit request on 4 December 2022
This edit request to Variants of SARS-CoV-2 has been answered. Set the |answered= or |ans= parameter to no to reactivate your request. |
Alpha (VOC 20DEC-01), Beta, and COH.20G/501Y (identified in Columbus, Ohio).[1][1] This last became the dominant form of the virus in Columbus in late December 2020 and January and appears to have evolved independently of other variants.[253][254].[2][3]
THE TWO SENTENCES ABOVE ARE WRONG AND SHOULD BE REMOVED FROM WIKIPEDIA. OHIO STATE UNIVERSITY HAS REMOVED THIS INFORMATION FROM ITS WEBPAGE. YOU CAN CHECK IT YOURSELF. IT IS FROM A PREPRINT, NOT A PUBLICATION. THESE TWO SENTENCES CONTAIN WRONG INFORMATION. 66.213.101.107 (talk) 19:42, 4 December 2022 (UTC)
References
- ^ Shahhosseini N, Babuadze GG, Wong G, Kobinger GP (April 2021). "Mutation Signatures and In Silico Docking of Novel SARS-CoV-2 Variants of Concern". Microorganisms. 9 (5): 926. doi:10.3390/microorganisms9050926. PMC 8146828. PMID 33925854. S2CID 233460887.
- ^ "Researchers Discover New Variant of COVID-19 Virus in Columbus, Ohio". wexnermedical.osu.edu. 13 January 2021. Archived from the original on 15 January 2021. Retrieved 16 January 2021.
- ^ Tu H, Avenarius MR, Kubatko L, Hunt M, Pan X, Ru P, et al. (26 January 2021). "Distinct Patterns of Emergence of SARS-CoV-2 Spike Variants including N501Y in Clinical Samples in Columbus Ohio". bioRxiv 10.1101/2021.01.12.426407.
- Could you please clarify? I'm of a mind to remove the second sentence as not being sourced to WP:MEDRS standards, but the first citation seems kosher as published in a reputable journal (although it doesn't mention the "alpha" and "beta" terminology, so perhaps that should be removed). Ovinus (talk) 21:24, 9 December 2022 (UTC)
 Not done for now: please establish a consensus for this alteration before using the
Not done for now: please establish a consensus for this alteration before using the {{Edit semi-protected}}template. Also, please read policies like WP:RS and explain which citation(s) are demonstrating that the information is wrong. Among Us for POTUS (talk) 06:40, 13 December 2022 (UTC)
Remove overview table?
Currently none of the trackers list any variant of concern beyond Omicron, so I think that table is now obsolete, does anybody disagree? Ideally, its contents should be moved to prose. Fernando Trebien (talk) 20:02, 8 December 2022 (UTC)
- agree -remove--Ozzie10aaaa (talk) 22:07, 9 December 2022 (UTC)
- Inclined to move it, as a valuable record of the history of the evolution of variants and reactions to them, perhaps to a new sub-subsection within Variants of concern (WHO) - maybe something like 'Summary of historic VoCs' - Yadsalohcin (talk) 09:25, 30 December 2022 (UTC)
Arcturus variant
Here are some sources on the variant also known as XBB.1.16:
- Arcturus: What we know about the new Covid variant and its symptoms, independent.co.uk
- New COVID-19 subvariant referred to as Arcturus on the rise in Australia. This is what we know about it, abc,net.au
Autarch (talk) 22:02, 4 May 2023 (UTC)
- thank you, will look--Ozzie10aaaa (talk) 23:00, 4 May 2023 (UTC)
- An article on the colloquial nomenclature of the variants is called for. Some are stars à la Arcturus (https://www.theguardian.com/world/2023/may/09/covid-variant-arcturus-conjunctivitis (as in the names of actual stars and/or wide media usage)), Kraken is a mythical beast (https://www.dictionary.com/e/new-covid-variant-name-list/), and some, like Nelly, Pelican, Quail, Mockingbird, and Eeek (https://www.theguardian.com/world/2021/feb/17/nicknames-emerge-for-growing-list-of-covid-variants) are for ease of discussion in the lab. And then there's Cerberus, Typhon, and Gryphon (more myths, which have been proposed (unless that's Typhoon)) (https://time.com/6229947/how-are-covid-19-variants-named/), etc. There is a lot to parse here (https://www.inc.com/rebecca-deczynski/covid-variant-names-branding-lessons.html)... Nomenclature is tricky subject. Nomenclature matters. kencf0618 (talk) 14:10, 24 May 2023 (UTC)
- Wrote it. Have at it. Colloquial names of COVID-19 variants kencf0618 (talk) 21:54, 24 May 2023 (UTC)
Needs updating for EG.5 / Eris
Recently noted as the dominant strain in US/UK infections - Keith D. Tyler ¶ 07:04, 8 August 2023 (UTC) https://www.cbsnews.com/news/covid-variant-eg-5-now-eris/?ftag=CNM-00-10aac3a
- will look at it--Ozzie10aaaa (talk) 16:04, 8 August 2023 (UTC)
BA.2.86
WHO has classified BA.2.86 as a “variant under monitoring”. https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-g20-health-ministers-meeting---inaugural-session---18-august-2023 76.92.198.127 (talk) 13:34, 29 August 2023 (UTC)
JN.1 Variant
It has recently popped up, and I am not sure if now would be a good idea to add it, or we wait for a bit of time until it's credible enough to add reliable sources. This has been something new since last year, thanks to the COVID vaccine being efficient in stopping the virus. lmk when this could be added. Yours truly, 🄼🄾🄳 🄲🅁🄴🄰🅃🄾🅁 (talk) 03:11, 12 December 2023 (UTC)
- Seems to be causing enough alarm that Singapore and Indonesia are introducing (minor) restrictions, so we should add it: https://www.livemint.com/news/world/singapore-government-implements-preventive-norms-for-travellers-amid-covid-scare-11702518104782.html Jpatokal (talk) 21:08, 14 December 2023 (UTC)
Incoming redirect JN.1
This edit request has been answered. Set the |answered= or |ans= parameter to no to reactivate your request. |
Please add a hatnote to handle incoming redirect JN.1 (edit | talk | history | protect | delete | links | watch | logs | views)
Please add
{{redirect-multi|1|JN.1|other uses|JN1 (disambiguation)}}
-- 65.92.247.66 (talk) 11:44, 23 December 2023 (UTC)
- thanks, done, added under 'Further sublineages emerging in 2023' Yadsalohcin (talk) 12:47, 23 December 2023 (UTC)
Currently Circulating Variant of Concern
please readd the "Delta variant" to the currently circulating variant of concern. 119.94.169.2 (talk) 07:32, 3 January 2024 (UTC)
JN.1 - split off alone or as section of BA.2.86?
Sooner or later it might make sense to either split off JN.1 into its own article, or as a subsection of BA.2.86. After all, it's the overwhelmingly dominant variant in four of the six regions of the world.[1] Boud (talk) 15:01, 10 February 2024 (UTC)
References
- ^ Updated Risk Evaluation of JN.1, 09 January 2023 [mislabelled date] (PDF), World Health Organization, 9 February 2024, Wikidata Q124477897, archived (PDF) from the original on 10 February 2024
WHO citation "Tracking SARS-CoV-2 variants" [4]
In the main section of the article, citation 4 is listed as "failed verification" because its most recent listed accessed date is 2022 and the sentence citing the page lists a date in 2024. This WHO page (link) is updated frequently. At time of writing, the apparent most recent update was 15 April 2024, when an updated risk evaluation for JN.1 was added, and the page itself claims to be up to date as of 3 May 2024. What is the appropriate way to cite a single URL which updates frequently? 184.62.88.78 (talk) 20:24, 9 May 2024 (UTC)
- will look--Ozzie10aaaa (talk) 22:44, 9 May 2024 (UTC)
- Have a look at Tracking SARS-CoV-2 variants, Wikidata Q127328768 – which is a generic element with multiple versions. Currently it has two specific versions:
- Tracking SARS-CoV-2 variants, World Health Organization, 20 December 2023, Wikidata Q127328911, archived from the original on 20 December 2023
- Tracking SARS-CoV-2 variants, World Health Organization, 28 June 2024, Wikidata Q127328784, archived from the original on 10 July 2024.
- See {{cite Q}} for documentation on using cite Q. The generic Wikidata property has edition or translation (P747) should be added to the generic element such as Q127328768, and the property edition or translation of (P629) should be added to the specific element representing a source of the page, such as Q127328911 or Q127328784. Boud (talk) 17:29, 10 July 2024 (UTC)
- Have a look at Tracking SARS-CoV-2 variants, Wikidata Q127328768 – which is a generic element with multiple versions. Currently it has two specific versions:
FLiRT
I'm somewhat reluctant to start a new section, but my source mentions this variant and doesn't say it began in 2023. If it didn't start in 2023, it must have started in 2024.— Vchimpanzee • talk • contributions • 19:06, 14 May 2024 (UTC)
- yes, 2024--Ozzie10aaaa (talk) 18:14, 10 July 2024 (UTC)
- This is in a 2024 section now. Dan Bloch (talk) 18:32, 10 July 2024 (UTC)
Omicron in lead section
The sentence in lead section "As of June 2024, only Omicron is designated as a circulating variant of concern by the World Health Organization." is incorrect. The website itself (updated on 5 June 2024) used as a source for this sentence doesn't list any circulating VoCs.
This WHO's statement from 16 March 2023 states "With these changes factored in, Alpha, Beta, Gamma, Delta as well as the Omicron parent lineage (B.1.1.529) are considered previously circulating VOCs."
Omicron's sublineages and their descendants were given separate labels such as VOI and VUM. KapSoule (talk) 11:33, 16 June 2024 (UTC)
 Fixed Boud (talk) 17:18, 10 July 2024 (UTC)
Fixed Boud (talk) 17:18, 10 July 2024 (UTC)
Tang paper / Retraction watch
Retraction watch has flagged an erratum re the Tang paper. The authors write: "We now recognize that within the context of our study the term “aggressive” is misleading and should be replaced by a more precise term “a higher frequency”. In short, while we have shown that the two lineages naturally co-exist, we provided no evidence supporting any epidemiological conclusion regarding the virulence or pathogenicity of SARS-CoV-2..." My impression is that the elements of the retraction don't affect the words currently used in this article. Could this be checked by an expert? Yadsalohcin (talk) 08:44, 14 June 2024 (UTC)
- thank you for post--Ozzie10aaaa (talk) 18:13, 10 July 2024 (UTC)
Historical variant sections need re-factoring
As of the current version of 18:17, 10 July 2024, we have three sections that are useful (especially since there is no point having all the descendants of Omicron listed in SARS-CoV-2 Omicron variant, and nobody seems to be trying to add them there anyway), but don't quite make sense in terms of the overall structure of the article. They should be shifted to be either earlier or later than the three VOC, VOI, and VUM stand-alone sections, or be integrated into a single section:
- Variants of SARS-CoV-2#Overview of historical variants of concern or under monitoring
- Variants of SARS-CoV-2#Variant of concern lineages under monitoring (WHO) - which has a good table
- Variants of SARS-CoV-2#Previously circulating and formerly monitored variants (WHO)
I'm not sure how these should be re-organised, but it seems to me that the main principle should be that we don't want a particular variant to be listed multiple times in detail in these sections - because then people won't know which entry is the main one to read or edit.
My initial proposal is that integrating Variants of SARS-CoV-2#Overview of historical variants of concern or under monitoring and Variants of SARS-CoV-2#Variant of concern lineages under monitoring (WHO) into the three respective sections of Variants of SARS-CoV-2#Previously circulating and formerly monitored variants (WHO) would make the most sense, possibly addding one or two extra subsections e.g. for a table.
This is mainly a problem of lack of media attention to SARS-CoV-2 since 2021-ish: less media attention leads to less Wikipedia editing.
Please give any arguments for or against this proposal, or describe alternative proposals on how to reorganise these sections. Boud (talk) 18:33, 10 July 2024 (UTC)
- will look--Ozzie10aaaa (talk) 13:45, 11 August 2024 (UTC)
Merge proposal for SARS-CoV-2 variant of concern
- The following discussion is closed. Please do not modify it. Subsequent comments should be made in a new section. A summary of the conclusions reached follows.
- No comments. Merging.⸺(Random)staplers 06:33, 29 September 2024 (UTC)
With the latest variants not being "variants of concern", and with the transclusion of SARS-CoV-2 variant of concern taking up two-thirds of the lead, which is effectively most of the article, given the rest of the article is outdated or unnecessary,
- I propose merging SARS-CoV-2 variant of concern into Variants of SARS-CoV-2 under a "Definitions" section.⸺(Random)staplers 15:38, 10 September 2024 (UTC)
I also just realized the section "Notability Criteria" is just a copy of "Criteria". That section was copied into SARS-CoV-2 variant of concern, so attribution should not be necessary (for that section) if this merge goes through.⸺(Random)staplers 15:50, 10 September 2024 (UTC)