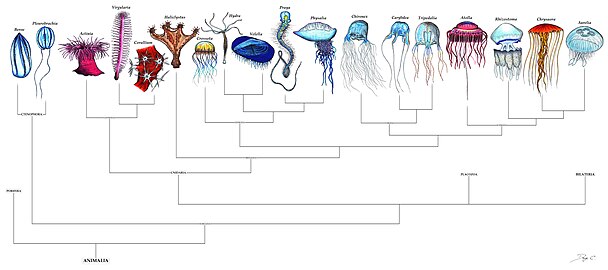
User:Epipelagic/sandbox/current1
| This is a Wikipedia user page. This is not an encyclopedia article or the talk page for an encyclopedia article. If you find this page on any site other than Wikipedia, you are viewing a mirror site. Be aware that the page may be outdated and that the user in whose space this page is located may have no personal affiliation with any site other than Wikipedia. The original page is located at https://en.wikipedia.org/wiki/User:Epipelagic/sandbox/current1. |
RESOURCES AND WORKING DRAFTS ONLY
Viruses in the ocean conveyor belt
[edit]Viruses shape marine ecosystems by controlling their host abundance and diversity through cell lysis and generate and maintain diversity through horizontal gene transfer (Angly et al., 2006; Rohwer and Thurber, 2009; Zeng and Chisholm, 2012). Moreover, viral lysis influences oceanic productivity by promoting organic matter and nutrient cycling through the release of intracellular material from the host cells (Middelboe et al., 1996; Middelboe and Lyck, 2002).[1]
Several parameters affect the distribution of viruses in the marine environment (De Corte et al., 2012, 2016; Brum et al., 2015). The presence of a suitable host is key to perpetuate the viral “life cycle.” However, other variables, such as temperature (Parada et al., 2007), UV radiation (Wommack et al., 1996) and salinity (Kukkaro and Bamford, 2009), influence the time span that viruses remain active in the ambient water and capable of infecting a host. The effects of these biotic and abiotic factors have been studied in surface waters (Bongiorni et al., 2005; De Corte et al., 2011) and, infrequently, in the deep ocean, where low temperature leads to relatively slower virus decay rates than in surface waters (Parada et al., 2007). The longer bacterial generation time and slower viral decay rates in the deep ocean (Parada et al., 2007; De Corte et al., 2012), together with the limited vertical mixing of the deep-water masses, suggest that oceanic currents and the thermohaline circulation may play an important role in influencing the global distribution of viruses and their hosts (McGillicuddy et al., 2007; Sintes et al., 2013, 2015; Sul et al., 2013; Brum et al., 2015; Frank et al., 2016).[1]
The global overturning circulation (Figure 1) is a system of oceanic currents driven by wind, density and mixing processes (Schmittner et al., 2007; Talley, 2013). The North Atlantic Deep Water (NADW) is formed in the North Atlantic Ocean through the sinking of cold and dense waters, and moves southward toward the Southern Ocean (Talley, 2013). The second limb of the deep overturning circulation system is the Antarctic Bottom Water (AABW). The AABW is formed in the Southern Ocean via mixing of NADW, Indian Deep Water, Pacific Deep Water, and shelf waters originating from Antarctic shelf systems (Talley, 2013).[1]
Viruses, as part of the dissolved organic matter pool (DOM; operationally defined as the organic matter that passes through a 0.2 μm filter), should behave conservatively and reflect the different formation sites of the deep waters (Bercovici and Hansell, 2016). However, it is important to note that certain local effects, such as particle export and in situ production (Nagata et al., 2000; Hansell and Ducklow, 2003; Yokokawa et al., 2013), play an important role in determining bacterial and viral abundance and community composition. Sinking particles provide a source of fresh substrate for microbes in the deep sea (Cho and Azam, 1988; Karl et al., 1988; Follett et al., 2014). Hence, this organic matter might be especially important in the Pacific Deep Water of the North Pacific with its low DOM concentration (Hansell, 2013).[1]
DNA viruses lack conserved marker genes, hindering the assessment of their biogeographical patterns. The dispersal of viruses across the oceans has only been recently explored (Angly et al., 2006; Hurwitz and Sullivan, 2013; Martinez-Hernandez et al., 2017; Gregory et al., 2019). Moreover, most studies addressing viral community composition are limited to the upper ocean layers (Angly et al., 2006; Brum et al., 2015; Hurwitz et al., 2015). Due to methodological limitations, virus-host interactions in the dark ocean have mainly been assessed by microbial and viral enumeration (De Corte et al., 2012; Wigington et al., 2016; Lara et al., 2017). Therefore, little is known about the changes of viral communities along the deep ocean conveyor belt circulation system.[1]
Although viromics (metagenomics of viral communities) is increasingly used to study viral diversity and function in the marine ecosystems (Breitbart et al., 2004; Breitbart and Rohwer, 2005; Hurwitz and Sullivan, 2013; Brum et al., 2015), it has its limitations. Viromics typically requires a large sample volume, which may limit the number of samples that can be collected and processed in a timely manner. Moreover, filtration of water samples to physically separate the viral fraction from other microorganisms may lead to the loss of a fraction of the viral community, in particular of giant viruses, while some small bacteria might pass through the filter (Martinez Martinez et al., 2014).[1]
References
[edit]- ^ a b c d e f De Corte, Daniele; Martínez, Joaquín Martínez; Cretoiu, Mariana Silvia; Takaki, Yoshihiro; Nunoura, Takuro; Sintes, Eva; Herndl, Gerhard J.; Yokokawa, Taichi (2019-08-21). "Viral Communities in the Global Deep Ocean Conveyor Belt Assessed by Targeted Viromics". Frontiers in Microbiology. 10. Frontiers Media SA. doi:10.3389/fmicb.2019.01801. ISSN 1664-302X.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Modified material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Microbial biogeochemical roles and ecosystem functioning
[edit]
Phylogeny and functional traits
[edit]To better understand microbial ecosystem functioning, creating links between an individual taxon and a specific function is required. However, it is infeasible to trace all the diverse biogeochemical processes and to relate them to taxa. This problem may be solved from the point of view of the microorganisms, as microbial functional capabilities have been found to connect strongly with phylogeny (Martiny et al. 2013; Zimmerman et al. 2013). For example, the presence of functional genes related to oxygenic photosynthesis, methane oxidation and sulfate reduction has been found to be highly phylogenetically conserved (Martiny et al. 2013). In coastal seawater, recent studies have shown that microorganisms that assimilated organic matter, including starch and glucose, were phylogenetically clustered (Bryson et al. 2017; Mayali and Weber 2018), reflecting phylogenetically conserved resource partitioning in the coastal microbial loop (Bryson et al. 2017). Such phylogenetic conservation in substrate utilization supports similar distribution patterns (even at a broader taxonomic level; Philippot et al. 2010; Schmidt et al. 2016) and similar lifestyles among microbial relatives. Salazar et al. (2015) found that the particle-associated and free-living populations in the deep ocean had different phylogenetic origins. These observations enhance the possibility of inferring microbial functional traits with phylogenetic information.[1]
However, growing evidence now suggests that variations in functional traits occur within closely related microbes, even at the loosely defined species level (Larkin and Martiny 2017). Prochlorococcus, the most abundant genus of photosynthetic organisms, diverges into high- and low-light-adapted ecotypes, which display different light-harvesting strategies (Bibby et al. 2003). Likewise, Alteromonas macleodii, a typical copiotrophic r-strategist, contains both surface and deep-sea ecotypes (Ivars-Martinez et al. 2008), which have been shown to differ substantially in their capacity to degrade algal polysaccharides (Neumann et al. 2015). On the other hand, microorganisms performing similar metabolic functions can be only distantly related (Martiny et al. 2015), the basic principle of functional redundancy. Louca et al. (2016, 2017) found high functional redundancy in both marine and plant-associated microbial communities, implying that microbial functional traits are widely spread among microbial lineages.[1]
Horizontal gene transfer and gene gain and loss likely result in the discrepancy in phylogeny-functional trait relationships (Fig. 2a). The type of functional trait is also an important contributor, with traits with slower evolutionary rates displaying clearer phylogenetically conserved patterns. Bryson et al. (2017) reported that traits of resource utilization in marine microbes, represented by patterns of substrate assimilation, were phylogenetically cohesive, whereas those of biosynthetic activity after assimilation were heterogeneous. Additionally, functional potential (gene presence) does not necessarily mean function execution (gene expression) and functional capability is not equal to rate (the same functional trait can be different in rate). Although the capability of glucose incorporation is widespread in bacteria and displays a shallow phylogenetic clustering (Martiny et al. 2013), the glucose assimilation rates in soil bacteria have been observed to vary greatly across phylogeny and display a deep phylogenetic clustering (Morrissey et al. 2016). This finding highlights the importance of incorporating functional rates into phylogeny-function relatedness and raises the need to examine how quantitative changes in microbial function affect ecosystem functioning.[1]
Diversity and ecosystem functioning
[edit]There is growing evidence of a positive relationship between microbial diversity and ecosystem functioning (Cardinale et al. 2012; Delgado-Baquerizo et al. 2016b; Schnyder et al. 2018), although negative or no relationships have also been reported (Becker et al. 2012). Such relationships are derived primarily from studies on the terrestrial environment and have rarely been assessed for marine microbial communities. A study of microbial diversity-ecosystem functioning (DEF) relationship in marine surface water also supported a positive correlation, showing that a more phylogenetically diverse bacterial community had a greater level of ecosystem functioning (heterotrophic productivity measured by leucine incorporation; Galand et al. 2015). The enhancement of ecosystem functioning by increased biodiversity is thought to result from complementarity (minimal overlap) in resource use by functionally distinct taxa (Petchey and Gaston 2002) and/or through inter taxa facilitation (Hooper et al. 2005). Therefore, the relationship between diversity and ecosystem functioning is controlled by the niche-based mechanisms: differentiation in resource niche and selection effect (Krause et al. 2014).[1]
The few studies that investigated the shape of the positive relationship between microbial diversity and ecosystem functioning have frequently uncovered a more linear relationship (Delgado-Baquerizo et al. 2016a) than the approaching-flat relationship seen for plants and animals (Cardinale et al. 2011). Such a linear relationship implies an indefinite increase of ecosystem functioning with increasing microbial diversity, challenging the idea of functional redundancy as mentioned above. In fact, Galand et al. (2018) provide evidence against the hypothesis of functional redundancy by showing a strong link between marine microbial community compositions and functional attributes using all the set of metagenomic reads. The authors emphasize the need to consider all functional aspects rather than relying only on known genes in investigating microbial DEF relationships. In addition, different processing rates seen in the same functional trait (Morrissey et al. 2016) may also provide opposing evidence against functional redundancy. However, these findings do not rule out the possibility for a partial functional redundancy, implicating that different types of functional traits may have different levels of redundancy. The idea of functional redundancy on the one hand can help to explain the high level of marine microbial diversity (different taxa are supported by a limited range of resources and conduct the same set of metabolic processes; Allison and Martiny 2008), while on the other hand can limit the extent of ecosystem functioning.[1]
Diversity is composed of different components, including richness (taxonomic diversity), phylogeny (phylogenetic diversity) and function (functional diversity) (Fig. 2b). Different types of diversity can inform distinct microbial DEF relationships. However, taxonomic diversity is the more frequently used proxy in inferring DEF relationships, compared to functional diversity and phylogenetic diversity. It has been reported that taxonomic diversity has relatively little impact on ecosystem functioning (Nielsen et al. 2011), while functional diversity was more correlated, mostly likely by determining ecological niches and inter taxa interactions (Hooper et al. 2005; Krause et al. 2014). Nevertheless, functional diversity is always difficult to measure (functional activity) and/or requires additional sequencing efforts to analyze (functional genes). Thus, phylogenetic diversity is increasingly implemented as a proxy of functional diversity, with the thought that many functional traits are phylogenetically conserved. Indeed, a positive correlation has been found between marine surface bacterial productivity and phylogenetic diversity of the active community; no similar association was found when taxonomic diversity (Shannon index) was analyzed (Galand et al. 2015). The findings of Galand et al. (2015) highlighted that ecosystem functioning is related to the active rather than the total community that contains dormant taxa. This provides an explanation for the more frequently observed negative and/or no relationships between phylogenetic diversity of the total community and ecosystem functioning (Goberna and Verdu 2018; Pérez-Valera et al. 2015; Severin et al. 2013). The relationship between phylogenetic diversity and ecosystem functioning also relates to taxon-specific functional capability and evolutionary history (Gravel et al. 2012). Under which conditions phylogenetic diversity can be used as a representative of functional diversity should be characterized further.[1]
The microbial DEF relationship can be confounded by environmental variations, as environmental factors can exert influences on both diversity and ecosystem functioning. Orland et al. (2018) demonstrated that pH and organic matter quantity and quality explained as much variation in CO2 production as did taxonomic diversity in lake sediments; these environmental factors exerted direct influences on ecosystem functioning due to their unrelatedness to taxonomic diversity. Comparatively, Delgado-Baquerizo et al. (2016b) found in a global set of soil samples that the DEF relationship was maintained when accounting for edaphic factors, which suggests that taxonomic diversity can exert influences on ecosystem functioning independently of environmental variabilities. A global survey of microbiome in seawater showed a decoupling of taxonomy and function, with the latter being more susceptible to environmental changes (Louca et al. 2016). Environmental conditions determine the availability of electronic donors/acceptors to microbes and shape the process of biogeochemical reactions. In addition to the environment, stochastic processes may also affect diversity and influence ecosystem functioning (Orland et al. 2018; Zhou et al. 2013). Overall, the positive DEF relationship in microorganisms is supportive of phylogenetic conservation in functional traits. More effort is needed to discern the role of different diversity components and the role of deterministic and stochastic processes in determining ecosystem functioning.[1]
Ecosystem functioning prediction
[edit]The abovementioned relationships have stimulated great interest in using microbial data to predict ecosystem functioning (statistical simulation instead of direct measurement) (Graham et al. 2016; Powell et al. 2015b) (Fig. 2b). Here, we focus on the interactions between community (diversity and abundance) and ecosystem functioning, although physiological properties can also be related (Wieder et al. 2013). Graham et al. (2016) synthesized 82 global datasets from different ecosystems to improve the predictive power of carbon and nitrogen processing rates by the inclusion of the microbial community data. They found that the addition of both compositional and diversity data could strengthen the predictive power, although this was not applicable to all datasets. Andersson et al. (2014) demonstrated, via structural equation models, that the model that included total bacterial abundance explained 54% of the variation in nitrogenase activity in coastal sediments and by replacing total bacterial abundance with cyanobacterial biomass it could increase the predictive power.[1]
The explanatory power of microbial data in predictive models is always lower than that of abiotic factors (Graham et al. 2014, 2016; Powell et al. 2015b), consistent with the notion that environmental factors have direct impacts on ecosystem functioning. Moreover, under different environmental conditions (e.g., temperature, Dolan et al. 2017), the explanatory power of microbial data may change. However, this does not decrease the importance of microbial data in functional prediction. Recently, Zhang et al. (2018) found that in coastal sediments adding different copy number ratios of functional and rRNA genes into stepwise regression models substantially increased the predictive power of denitrification and anammox rates, although alpha diversity and gene abundance of involved bacteria were poorly correlated to the function potentials. In addition to abundance and diversity, it is also important to include microbial interactions to the predictive model in future studies (Fig. 2b). In summary, knowledge on the distribution patterns of microbial communities is indispensable for understanding their biogeochemical and ecological functions.[1]
References
[edit]- ^ a b c d e f g h i j Liu, Jiwen; Meng, Zhe; Liu, Xiaoyue; Zhang, Xiao-Hua (2019). "Microbial assembly, interaction, functioning, activity and diversification: A review derived from community compositional data". Marine Life Science & Technology. 1 (1): 112–128. Bibcode:2019MLST....1..112L. doi:10.1007/s42995-019-00004-3. S2CID 208569970.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Inference of microbial activity
[edit]
As stated above, a microbial community contains both active and dormant members. Dormant microbes constitute the seed bank of a community. Their repeated entrance to and exit from the seed bank decouple the active and total fractions (Jones and Lennon 2010; Fig. 3). Since RNA is a good indicator of whether or not a microbe is viable, rRNA based methods have been extensively used to characterize the active community, despite the difficulty in RNA extraction and the probable biases introduced by reverse transcription.[1]
Mounting evidence suggests that there is a significant difference between the total (resident) and active fractions within a microbial community. Several abundant taxa are less active in the RNA pool, whereas some highly active taxa show low abundance or are almost absent in the DNA pool (Baldrian et al. 2012; Richa et al. 2017; Romanowicz et al. 2016; Sebastián et al. 2018). For example, Cyanobacteria and the SAR11 clade of Alphaproteobacteria are the most abundant microbes in the global surface ocean; whereas the former is always disproportionately active (16S rRNA:rDNA > 1), the latter tends to be less active (Campbell and Kirchman 2013; Hunt et al. 2013; Zhang et al. 2014). Further, a refined phylogenetic analysis showed that different ecotypes of the SAR11 clade varied in their 16S rRNA:rDNA ratios (Salter et al. 2015). A similar phenomenon was also shown for different ecotypes of MG-I (Hugoni et al. 2013). Within a microbial community, activity can also vary between rare and abundant taxa (Campbell et al. 2011; Richa et al. 2017). Richa et al. (2017) reported that more than 70% of the rare taxa in coastal seawater of the Mediterranean Sea had high 16S rRNA:rDNA ratios. To explain this decoupling of abundance and activity, Campbell et al. (2011) proposed that a substantial proportion of bacteria became active when their abundance decreased, indicating that high abundance may be a constraint factor for activity or that top-down processes i.e., grazing and virus lysis could stimulate activity. Seasonality (Hugoni et al. 2013), environmental factors, such as salinity (Campbell and Kirchman 2013), and lifestyle modes (free-living and particle-attached; Li et al. 2018) have also been reported as drivers for 16S rRNA:rDNA ratio variations.[1]
The active and total microbial communities have been found to display contrasting biogeographic patterns and respond differently to environmental factors in seawater (Zhang et al. 2014). Environmental changes would generate uncomfortable conditions for active microbes, and the adaptation and successful establishment of active microbes in a new environment are difficult (Hanson et al. 2012). By contrast, the growth of several dormant microbes can be stimulated by changing environments, contributing to microbial community succession. Considering this, the active community is likely to display a stronger distance-decay relationship than the total community (Zhang et al. 2014). Nevertheless, our understanding of the assembling processes (relative role of deterministic and stochastic processes) of the total and active communities is limited. Zhang et al. (2014) also found that the active (dominated by negative correlations) and total (dominated by positive correlations) bacterial communities exhibited different co-occurrence patterns. Frequent transitions of microbes between an active or dormant status under changing environments would lead to variations in co-occurrence patterns, causing significant alterations in ecosystem functioning (Fig. 3). As mentioned above, active microbes are directly linked to ecosystem functioning by actively carrying out biogeochemical reactions (Nannipieri et al. 2003). Thus, distinguishing the active fraction from the whole community would provide novel insights into patterns of microbial assembly, co-occurrence and DEF relationship. Moreover, the finding of a higher environmental sensitivity of active heterotrophs than active autotrophs in seawater (Zhang et al. 2014) further raises the need to treat functionally different microbial groups separately.[1]
Another possible utilization of the rRNA:rDNA ratio is to indicate potential growth rate (Campbell et al. 2011; Lankiewicz et al. 2016), as numerous microorganisms are yet to be cultivated and their growth rates are not able to be measured (Kirchman 2016). A high rRNA:rDNA ratio may imply a high growth rate. Thus, the lower proportion of SAR11 in the RNA than in the DNA pool, as mentioned above, may indicate a slow growth mode, although it may also be due to the low number of ribosomes per cell, given its small cell size. The slow growth rate of SAR11, however, is further verified by culture-based analyses (Lankiewicz et al. 2016). In comparison, copiotrophic taxa from Alteromonas and the Rosebacter clade often display higher growth rates (Hunt et al. 2013; Lankiewicz et al. 2016). Noticeably, the rRNA:rDNA ratio has been reported to be not always effective in quantifying microbial growth rates, and protein synthesis potential has been proposed to be a more suitable interpretation (Blazewicz et al. 2013).[1]
References
[edit]- ^ a b c d e Liu, Jiwen; Meng, Zhe; Liu, Xiaoyue; Zhang, Xiao-Hua (2019). "Microbial assembly, interaction, functioning, activity and diversification: A review derived from community compositional data". Marine Life Science & Technology. 1 (1): 112–128. Bibcode:2019MLST....1..112L. doi:10.1007/s42995-019-00004-3. S2CID 208569970.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Images
[edit]
of seagrass meadows
(B) PCA for molecular inventory was performed using the COI gene. Each PCA was implemented using Jaccard dissimilarity matrices. Points are individual sediment samples taken at a specific core and mesh size, while colored ellipses are 95% confidence intervals for each meadow. For COI, the Sainte Marguerite cluster contains samples from 2010 (open circles) and 2011 (closed circles). Colors of the ellipses are specific to each meadow and these colors were maintained across both PCAs.[1]
Endosphere
[edit]
Some microorganisms, such as endophytes, penetrate and occupy the plant internal tissues, forming the endospheric microbiome. The arbuscular mycorrhizal and other endophytic fungi are the dominant colonizers of the endosphere.[3] Bacteria, and to some degree archaea, are important members of endosphere communities. Some of these endophytic microbes interact with their host and provide obvious benefits to plants.[4][5][6] Unlike the rhizosphere and the rhizoplane, the endospheres harbor highly specific microbial communities. The root endophytic community can be very distinct from that of the adjacent soil community. In general, diversity of the endophytic community is lower than the diversity of the microbial community outside the plant.[7] The identity and diversity of the endophytic microbiome of above-and below-ground tissues may also differ within the plant.[8][3][2]
"The plant endosphere is colonized by complex microbial communities and microorganisms, which colonize the plant interior at least part of their lifetime and are termed endophytes. Their functions range from mutualism to pathogenicity. All plant organs and tissues are generally colonized by bacterial endophytes and their diversity and composition depend on the plant, the plant organ and its physiological conditions, the plant growth stage as well as on the environment."[9]
History of the concept
[edit]
In the 19th century, following Louis Pasteur, the accepted belief was that healthy plants showing no sign of diseases were free of microorganisms, and in particular were free of bacteria.(Compant et al., 2012).
"For a long time, the scientific community thought that plants that do not show symptoms of diseases are free of microorganisms, particularly from bacteria. There were few early reports on bacterial colonization of the plant endosphere (Galippe, 1887; Laurent, 1889); Bacteria occupying root nodules of leguminous plants, nowadays well known as rhizobia being responsible for fixing atmospheric nitrogen, were discovered by Martinus Willem Beijerinck in 1888 (Beijerinck, 1888). In the same year, Hellriegel and Wilfarth (1888) reported that leguminous plants are independent on mineral N, further indicating the importance of the N‐fixing symbiosis between plants and rhizobia."[10]
"Active research on the plant endosphere as a habitat for non‐pathogenic bacteria started in the 1990s, triggered by the increasing number of reports on the beneficial effects of plant growth‐promoting rhizobacteria (PGPR). The pioneering work of Johanna Döbereiner on specific bacteria which, like Herbaspirillum seropedicae, colonize the endosphere of sugarcane and fix nitrogen (Baldani et al., 1986; Boddey and Döbereiner, 1988) stimulated, given their importance for the Brazilian economy, further research on bacteria colonizing the plant endosphere. The increasing interest in studying microbial communities in the environment together with the development of molecular, cultivation‐independent tools to study their community structure also triggered research on endosphere microbiota."[10]
"In the early 1990s, definitions came up on the term ‘endophyte’, mostly referring to microorganisms that inhabit internal plant tissues, at least some time of their lifecycle, without causing apparent harm or disease to their host (Petrini, 1991; Wilson, 1995). Although this definition has been used in many studies and represents a pragmatic distinction between endophytes and pathogenic colonizers, it has been recently revised by Hardoim et al. (2015). A revised definition was needed due to the understanding obtained in the last years showing that pathogenicity or mutualism of microorganisms may depend on many factors including the plant genotype, the environment and the co‐colonizing microbiota (Brader et al., 2017). Therefore, a clear distinction between non‐pathogenic microorganisms (i.e. endophytes) and pathogens is often not feasible without detailed functional analysis. Also, functional assignment of endophytes studied purely by molecular tools, e.g. by microbiome analysis based only on phylogenetic markers, is usually not possible. Therefore, Hardoim et al. (2015) suggested that the term endophyte should refer to the habitat only and include all microorganisms, which for all or part of their lifetime colonize internal plant tissues. In the present review, the term endophyte refers to any microorganism that can colonize internal tissues of plants, including pathogens."[10]
"In the last decade, host‐associated microbiota have gained increasing attention, triggered by spectacular findings on the role of the human microbiome for human health, behaviour and well‐being. Already in 1994, Jefferson postulated that the evolutionary selection unit is not the macro‐organism (e.g. the plant) but the macro‐organism and all its associated microorganisms that act in concert as a holobiont (Jefferson, 1994). This hypothesis was further elaborated in the highly debated hologenome theory of evolution (Zilber‐Rosenberg and Rosenberg, 2008; Theis et al., 2016). The rhizosphere is considered as an important component of the plant holobiont, but endophytes have received increasing attention due to their intimate interaction with plants. Despite this increasing awareness of microbial life within plants, the endosphere is often recognized as one habitat without considering the variety of microenvironments and dynamics of microenvironment conditions. We aim here to review the multiple facets of the plant endosphere environment for bacterial colonization and life and pinpoint to the methodological limitations and knowledge gaps, which need to be addressed to further elucidate the role of bacterial endophytes in plant physiology and ecosystem functioning."[10]
Endophytes
[edit]Leaf endosphere
[edit]Root endosphere
[edit]Examples
[edit]Fungal diversity
[edit]
among different plant compartments

root endosphere and rhizosphere
The seed microbiome
[edit]
Root endosphere of the olive
[edit]
The cultivated olive is one of the oldest domesticated trees,[13] and has been shaped over millennia as one of the most important agro-ecosystems in the Mediterranean Basin.[14][15] Olive cultivation is threatened by several abiotic (such as soil erosion) and biotic (attacks from insects, nematodes and pathogenic microbes) constraints. Among relevant phytopathogens present in soil microbiota that affect olive health are Oomycota species such as Phytophthora species, as well as higher fungi such as Verticillium dahliae.[14][16][17][18] In addition to traditional microbiological menaces which affect olive crops, there are emerging diseases like olive quick decline syndrome caused by Xylella fastidiosa [19] and increases in pathogen and arthropod attacks as a consequence of changing from traditional olive cropping systems to high-density tree orchards. However, as of 2020 the impact of high-density olive groves on soil-borne diseases has yet to be studied.[20] Another menace is climate change, which is expected to affect the incidence and severity of olive diseases.[16] Finally, the reduction in the number of olive cultivars due to commercial (for example improved yield) or phytopathological (as tolerance to diseases) reasons, a trend observed in many areas, will eventually lessen olive genetic diversity. Factors like these may have have profound impact on the composition, structure and functioning of below ground microbial communities.[18][15]
Comprehensive knowledge of microbial communities associated with the olive root system, including the root endosphere and the rhizosphere soil, is needed to better understand their influence on the development, health and fitness of this tree. A priori, the vast majority of the olive-associated microbiota must be composed of microorganisms providing either neutral or positive effects to the host. Indeed, recent literature provides solid evidence that olive roots are a good reservoir of beneficial microorganisms, including effective biocontrol agents.[21][22][23] Among the beneficial components of the plant-associated microbiota, endophytic bacteria and fungi are of particular interest for developing novel biotechnological tools aimed at enhancing plant growth and controlling plant diseases. Moreover, microorganisms able to colonize and survive within the plant tissue can adapted to the specific microhabitat/niche where they can provide their beneficial effects.[24] Besides endophytes, beneficial components of tree root-associated microbiota colonizing the rhizoplane and rhizosphere soil can also directly promote plant growth as bio-fertilization and phyto-stimulation, or alleviate stress caused by abiotic (environmental pollutants, drought, salinity resistance) or biotic constraints.[18][15]
The olive root endosphere consists of quite different microbiota compared to that found in the olive rhizosphere, though the rhizosphere has higher alpha diversity (richness and evenness).[25][26] In a 2019 study by Fernández-González et al., the olive genotype appeared to be the main factor shaping below ground microbial communities, this factor being more determinant for the rhizosphere than for the endosphere, and more crucial for the bacteriota than for the mycobiota.[15] In this study, Proteobacteria (26% average relative abundance) was clearly dominated by Actinobacteria (64% average relative abundance) in the root endosphere (a similar finding has been reported in Agave species, particularly during the dry season [25]). However no sequences belonging to the kingdom Archaea were detected in the root endosphere in the Fernández-González study, in contrast to the results by Müller et al. who reported that Archaea was a major group in the olive phyllosphere.[27] The olive-associated microbiota harbors an important reservoir of beneficial microorganisms that can be used as plant growth promotion and/or biocontrol tools.[21][27] Moreover, bacterial antagonists of olive pathogens isolated from the olive root endosphere or the rhizosphere have the advantage to be adapted to the ecological niche where they can potentially exert their beneficial effect.[23][15] In the olive below ground (endophytic and rhizosphere) core bacteriota reported by Fernández-González, genera from which some species have been well characterized and described as biocontrol agents were present. For instance, Streptomyces was the second most abundant genus in the endosphere whereas Bacillus was the tenth more abundant in the rhizosphere. While Pseudomonas was part of the rhizosphere core bacteriota, it was not considered as constituent of the endophytic core because it was not always present in the root endosphere. Nevertheless, Pseudomonas was relatively much more abundant inside olive root tissues than in the rhizosphere. Regarding the core mycobiota, and as mentioned above, the most noticeable presence of a pathogenic fungus was Macrophomina, and to a lesser extent Colletotrichum.[15]
See also
[edit]- Endophyte → leaf endophyte / root endophyte
References
[edit]- ^ Cowart, Dominique A.; Pinheiro, Miguel; Mouchel, Olivier; Maguer, Marion; Grall, Jacques; Miné, Jacques; Arnaud-Haond, Sophie (2015). "Metabarcoding is Powerful yet Still Blind: A Comparative Analysis of Morphological and Molecular Surveys of Seagrass Communities". PLOS ONE. 10 (2): e0117562. Bibcode:2015PLoSO..1017562C. doi:10.1371/journal.pone.0117562. PMC 4323199. PMID 25668035.
- ^ a b Dastogeer, Khondoker M.G.; Tumpa, Farzana Haque; Sultana, Afruja; Akter, Mst Arjina; Chakraborty, Anindita (2020). "Plant microbiome–an account of the factors that shape community composition and diversity". Current Plant Biology. 23: 100161. doi:10.1016/j.cpb.2020.100161. S2CID 225626117.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Vokou, Despoina; Vareli, Katerina; Zarali, Ekaterini; Karamanoli, Katerina; Constantinidou, Helen-Isis A.; Monokrousos, Nikolaos; Halley, John M.; Sainis, Ioannis (2012). "Exploring Biodiversity in the Bacterial Community of the Mediterranean Phyllosphere and its Relationship with Airborne Bacteria". Microbial Ecology. 64 (3): 714–724. doi:10.1007/s00248-012-0053-7. PMID 22544345. S2CID 17291303.
- ^ Cite error: The named reference
Bulgarelli2012was invoked but never defined (see the help page). - ^ Dastogeer, Khondoker M.G.; Li, Hua; Sivasithamparam, Krishnapillai; Jones, Michael G.K.; Du, Xin; Ren, Yonglin; Wylie, Stephen J. (2017). "Metabolic responses of endophytic Nicotiana benthamiana plants experiencing water stress". Environmental and Experimental Botany. 143: 59–71. doi:10.1016/j.envexpbot.2017.08.008.
- ^ Rodriguez, R. J.; White Jr, J. F.; Arnold, A. E.; Redman, R. S. (2009). "Fungal endophytes: Diversity and functional roles". New Phytologist. 182 (2): 314–330. doi:10.1111/j.1469-8137.2009.02773.x. PMID 19236579.
- ^ Cite error: The named reference
Schlaeppi2014was invoked but never defined (see the help page). - ^ Abdelfattah, Ahmed; Wisniewski, Michael; Schena, Leonardo; Tack, Ayco J.M. (2020-05-14). "Experimental Evidence of Microbial Inheritance in Plants and Transmission Routes from Seed to Phyllosphere and Root". doi:10.21203/rs.3.rs-27656/v1.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Compant, Stéphane; Cambon, Marine C.; Vacher, Corinne; Mitter, Birgit; Samad, Abdul; Sessitsch, Angela (2021). "The plant endosphere world – bacterial life within plants". Environmental Microbiology. 23 (4): 1812–1829. doi:10.1111/1462-2920.15240. PMID 32955144. S2CID 221825778.
- ^ a b c d Compant, Stéphane; Cambon, Marine C.; Vacher, Corinne; Mitter, Birgit; Samad, Abdul; Sessitsch, Angela (2020). "The plant endosphere world – bacterial life within plants". Environmental Microbiology. 23 (4): 1812–1829. doi:10.1111/1462-2920.15240. PMID 32955144. S2CID 221825778.
- ^ a b Qian, Xin; Li, Hanzhou; Wang, Yonglong; Wu, Binwei; Wu, Mingsong; Chen, Liang; Li, Xingchun; Zhang, Ying; Wang, Xiangping; Shi, Miaomiao; Zheng, Yong; Guo, Liangdong; Zhang, Dianxiang (2019). "Leaf and Root Endospheres Harbor Lower Fungal Diversity and Less Complex Fungal Co-occurrence Patterns Than Rhizosphere". Frontiers in Microbiology. 10: 1015. doi:10.3389/fmicb.2019.01015. PMC 6521803. PMID 31143169.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Mitter, Birgit; Pfaffenbichler, Nikolaus; Flavell, Richard; Compant, Stéphane; Antonielli, Livio; Petric, Alexandra; Berninger, Teresa; Naveed, Muhammad; Sheibani-Tezerji, Raheleh; von Maltzahn, Geoffrey; Sessitsch, Angela (2017). "A New Approach to Modify Plant Microbiomes and Traits by Introducing Beneficial Bacteria at Flowering into Progeny Seeds". Frontiers in Microbiology. 8: 11. doi:10.3389/fmicb.2017.00011. PMC 5253360. PMID 28167932. S2CID 10675766.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Uylaşer, Vildan; Yildiz, Gökçen (2014). "The Historical Development and Nutritional Importance of Olive and Olive Oil Constituted an Important Part of the Mediterranean Diet". Critical Reviews in Food Science and Nutrition. 54 (8): 1092–1101. doi:10.1080/10408398.2011.626874. PMID 24499124. S2CID 26574218.
- ^ a b López-Escudero, Francisco Javier; Mercado-Blanco, Jesús (2011). "Verticillium wilt of olive: A case study to implement an integrated strategy to control a soil-borne pathogen". Plant and Soil. 344 (1–2): 1–50. doi:10.1007/s11104-010-0629-2. S2CID 13189260.
- ^ a b c d e f Fernández-González, Antonio J.; Villadas, Pablo J.; Gómez-Lama Cabanás, Carmen; Valverde-Corredor, Antonio; Belaj, Angjelina; Mercado-Blanco, Jesús; Fernández-López, Manuel (2019). "Defining the root endosphere and rhizosphere microbiomes from the World Olive Germplasm Collection". Scientific Reports. 9 (1): 20423. Bibcode:2019NatSR...920423F. doi:10.1038/s41598-019-56977-9. PMC 6938483. PMID 31892747.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Olive Diseases and Disorders. 2011. ISBN 9788178955391.
- ^ Sanzani, S.M.; Schena, L.; Nigro, F.; Sergeeva, V.; Ippolito, A.; Salerno, M.G. (2012). "Abiotic Diseases of Olive". Journal of Plant Pathology. 94 (3). doi:10.4454/JPP.FA.2012.069.
- ^ a b c Mercado-Blanco, Jesús; Abrantes, Isabel; Barra Caracciolo, Anna; Bevivino, Annamaria; Ciancio, Aurelio; Grenni, Paola; Hrynkiewicz, Katarzyna; Kredics, László; Proença, Diogo N. (2018). "Belowground Microbiota and the Health of Tree Crops". Frontiers in Microbiology. 9: 1006. doi:10.3389/fmicb.2018.01006. PMC 5996133. PMID 29922245.
- ^ Saponari, M.; Boscia, D.; Nigro, F.; Martelli, G.P. (2013). "Identification of DNA Sequences Related to Xylella Fastidiosa in Oleander, Almond and Olive Trees Exhibiting Leaf Scorch Symptoms in Apulia (Southern Italy)". Journal of Plant Pathology. 95 (3). doi:10.4454/JPP.V95I3.035.
- ^ Rallo, Luis; Barranco, Diego; Castro-García, Sergio; Connor, David J.; Gómez Del Campo, María; Rallo, Pilar (2014). "High-Density Olive Plantations". Horticultural Reviews Volume 41. pp. 303–384. doi:10.1002/9781118707418.ch07. ISBN 9781118707418.
- ^ a b Aranda, Sergio; Montes-Borrego, Miguel; Jiménez-Díaz, Rafael M.; Landa, Blanca B. (2011). "Microbial communities associated with the root system of wild olives (Olea europaea L. Subsp. Europaea var. Sylvestris) are good reservoirs of bacteria with antagonistic potential against Verticillium dahliae". Plant and Soil. 343 (1–2): 329–345. doi:10.1007/s11104-011-0721-2. S2CID 45997008.
- ^ Gómez-Lama Cabanás, Carmen; Legarda, Garikoitz; Ruano-Rosa, David; Pizarro-Tobías, Paloma; Valverde-Corredor, Antonio; Niqui, José L.; Triviño, Juan C.; Roca, Amalia; Mercado-Blanco, Jesús (2018). "Indigenous Pseudomonas SPP. Strains from the Olive (Olea europaea L.) Rhizosphere as Effective Biocontrol Agents against Verticillium dahliae: From the Host Roots to the Bacterial Genomes". Frontiers in Microbiology. 9: 277. doi:10.3389/fmicb.2018.00277. PMC 5829093. PMID 29527195. S2CID 3482295.
- ^ a b Gómez-Lama Cabanás, Carmen; Ruano-Rosa, David; Legarda, Garikoitz; Pizarro-Tobías, Paloma; Valverde-Corredor, Antonio; Triviño, Juan; Roca, Amalia; Mercado-Blanco, Jesús (2018). "Bacillales Members from the Olive Rhizosphere Are Effective Biological Control Agents against the Defoliating Pathotype of Verticillium dahliae". Agriculture. 8 (7): 90. doi:10.3390/agriculture8070090.
- ^ Mercado-Blanco, Jesus; Lugtenberg, Ben (2014). "Biotechnological Applications of Bacterial Endophytes". Current Biotechnology. 3: 60–75. doi:10.2174/22115501113026660038.
- ^ a b Coleman-Derr, Devin; Desgarennes, Damaris; Fonseca-Garcia, Citlali; Gross, Stephen; Clingenpeel, Scott; Woyke, Tanja; North, Gretchen; Visel, Axel; Partida-Martinez, Laila P.; Tringe, Susannah G. (2016). "Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species". New Phytologist. 209 (2): 798–811. doi:10.1111/nph.13697. PMC 5057366. PMID 26467257.
- ^ Beckers, Bram; Op De Beeck, Michiel; Weyens, Nele; Boerjan, Wout; Vangronsveld, Jaco (2017). "Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees". Microbiome. 5 (1): 25. doi:10.1186/s40168-017-0241-2. PMC 5324219. PMID 28231859.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Müller, Henry; Berg, Christian; Landa, Blanca B.; Auerbach, Anna; Moissl-Eichinger, Christine; Berg, Gabriele (2015). "Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees". Frontiers in Microbiology. 6: 138. doi:10.3389/fmicb.2015.00138. PMC 4347506. PMID 25784898.
Phyllosphere
[edit]
- references
- ^ Compant, Stéphane; Cambon, Marine C.; Vacher, Corinne; Mitter, Birgit; Samad, Abdul; Sessitsch, Angela (2020). "The plant endosphere world – bacterial life within plants". Environmental Microbiology. 23 (4): 1812–1829. doi:10.1111/1462-2920.15240. PMID 32955144. S2CID 221825778.
- ^ Qian, Xin; Chen, Liang; Guo, Xiaoming; He, Dan; Shi, Miaomiao; Zhang, Dianxiang (2018). "Shifts in community composition and co-occurrence patterns of phyllosphere fungi inhabiting Mussaenda shikokiana along an elevation gradient". PeerJ. 6: e5767. doi:10.7717/peerj.5767. PMC 6187995. PMID 30345176.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Seagrass meadows
[edit]Seagrasses are marine angiosperms that returned from the land to the marine environment 60–90 MYA in multiple events, resulting in a paraphyletic group composed of four families, three of which include only marine species.[1] Seagrasses are widely distributed in coastal waters all over the world (except Antarctica), from very shallow to 90 m depth.[2][3] Considered as ecosystem engineers,[4] seagrasses form extensive meadows, which are the foundation of complex systems. These meadows provide key ecosystem servicesm[3] i.e., natural processes or components that benefit human needs.[5] The three-dimensional structure of seagrass meadows provides critical habitats for many organisms and also serves as a nursery ground for juvenile stages of many species, including economically important species of finfish and shellfish.[6][7][8] In addition, seagrasses stabilize soft sediments and stock large quantities of CO2 as biomass and dead organic matter (blue carbon), contributing to the mitigation of anthropogenic emissions.[3][9][10][11]
Seagrass losses
[edit]Despite the ecological and economic importance of seagrass beds, an increasing number of reports have documented the ongoing loss of seagrass biomass in several countries, with a global decline rates estimated at 2–5% per year [11]. Most of these losses have been associated with human-driven pressures, including increased nutrient and sediment runoff, pollution, hydrological alterations, invasive species, commercial fishing or aquaculture practices. Not only do these pressures continue to threaten seagrass meadows, but they have also already shown to have caused large seagrass losses and regression worldwide, affecting entire ecosystems with a bottom up effect [12,13,14]. Despite the recent slowing down of these trends, mainly of fast-growing species [15], the overall worldwide trend is still the decline or deterioration. These ongoing declines are particularly high for the more slow-growing seagrasses (e.g., Posidonia oceanica; the foundation of one of the most valuable Mediterranean ecosystems), where reductions and a lack of recovery cause severe losses of some of the ecosystem services provided, since their ecological functions and services cannot be replaced by the fast growing seagrass species [5,16].[11]
To mitigate seagrass losses, it is essential to detect the environmental changes and seagrass stressors, prior to the decline (sometimes irreversibly) in local meadows occurs. The current challenge, thus, is the development and the combination of sensitive and measurable descriptors of seagrass stress, able to assess seagrass ecological state and its alterations. This will help to prevent the decline and enhance our understanding of seagrass global processes and threats, further supporting the development of effective monitoring and integrated management programs [17,18,19,20].[11]
The seagrass microbiome
[edit]To date, seagrass descriptors have focused on different biological organization levels from the population to the individual levels, such as shoot density, alongside biochemical and genetics descriptors [19,20]. The time of response of such indicators to stressors generally increases with the structural complexity, while their specificity decreases [18]. With most the descriptors used these days in seagrass monitoring programs being based on the slow responding population level (e.g., species composition, percent of cover, density, etc.), there is a growing interest in shortening the time of response by identifying early warning indicators. Recent studies [21,22,23,24,25,26,27,28,29,30,31,32] have evaluated the pivotal role of the microbial community associated with seagrass in their physiology and ecology, assessing their symbiotic relationships and the variability of microbes according to the environmental/host conditions, arguing that the host associated microbes could be a sensitive monitoring tool and ecological indicator. In fact, as microbial communities respond rapidly to environmental disturbance, monitoring their composition could represent an early indicator of environmental stress [21,22,24,25,29,31,32]. Despite the encouraging results of the above-mentioned studies, the comprehension of the seagrass associated microbial community variation according to the host conditions is still far from being clear.[11]
The seagrass-microbes associations are the result of a selective process involving both the seagrass microenvironment availability—depending on the host ecological/physiological conditions in response to the environment, and the metabolic capabilities of the microbes. The intimate relationship between the host and its associated microorganisms has led to considering them as a complex single super-organism that jointly responds to environmental changes as a functional complex unit, called the holobiont [33]. This perspective significantly changes the way of thinking of a living-organism and may provide important insights into the organism condition and, consequently, into the potential use of associated microbes as a source of ecological information.[11]
While several seagrass–microbe interactions, mainly studied as belowground functional processes, have been identified (see Section 1.2); a unified point of view about the factors implicated in the settling, composition and spatial–temporal variation of the associated microbial communities, is still missing. One of the main questions that need addressing is whether seagrass species-specificity can be hypothesized. In other words, does each seagrass species harbors its own specific taxonomic and/or functional microbiota across different sites, or is the seagrass microbiota shaped by the environmental conditions and follow a common pattern in different seagrass species growing in the same habitat. This is a pivotal point that would give insights into both the seagrass’ capability to select their hosts and the ecological meaning of variations in the microbial component of the seagrass holobiont: elucidating the key host–microbe interactions may provide guidance for seagrass managing and/or restoration [34].[11]
Often, different studies found heterogeneous results on the epiphytic microbial taxonomic composition; in some cases species-specific differences were found [35,36,37], while in others comparable microbial communities were found associated with different seagrass species from the same site [27,38,39]. The species-specificity of the microbial profile has been related to specific biochemical properties, while the comparable microbial communities were mainly attributed to the local environmental conditions.
- references
- ^ Hartog, C. den; Kuo, John (2006). "Taxonomy and Biogeography of Seagrasses". Seagrasses: Biology, Ecologyand Conservation. pp. 1–23. doi:10.1007/978-1-4020-2983-7_1. ISBN 978-1-4020-2942-4.
- ^ Duarte, Carlos M. (1991). "Seagrass depth limits". Aquatic Botany. 40 (4): 363–377. doi:10.1016/0304-3770(91)90081-F.
- ^ a b c Hemminga, Marten A.; Duarte, Carlos M. (19 October 2000). Seagrass Ecology. ISBN 9780521661843.
- ^ Wright, Justin P.; Jones, Clive G. (2006). "The Concept of Organisms as Ecosystem Engineers Ten Years on: Progress, Limitations, and Challenges". BioScience. 56 (3): 203. doi:10.1641/0006-3568(2006)056[0203:TCOOAE]2.0.CO;2. ISSN 0006-3568. S2CID 42248917.
- ^ Nordlund, Lina Mtwana; Koch, Evamaria W.; Barbier, Edward B.; Creed, Joel C. (2017). "Correction: Seagrass Ecosystem Services and Their Variability across Genera and Geographical Regions". PLOS ONE. 12 (1): e0169942. Bibcode:2017PLoSO..1269942N. doi:10.1371/journal.pone.0169942. PMC 5215874. PMID 28056075.
- ^ Beck, Michael W.; Heck, Kenneth L.; Able, Kenneth W.; Childers, Daniel L.; Eggleston, David B.; Gillanders, Bronwyn M.; Halpern, Benjamin; Hays, Cynthia G.; Hoshino, Kaho; Minello, Thomas J.; Orth, Robert J.; Sheridan, Peter F.; Weinstein, Michael P. (2001). "The Identification, Conservation, and Management of Estuarine and Marine Nurseries for Fish and Invertebrates". BioScience. 51 (8): 633. doi:10.1641/0006-3568(2001)051[0633:TICAMO]2.0.CO;2. ISSN 0006-3568. S2CID 27242795.
- ^ Jiang, Zhijian; Cui, Lijun; Liu, Songlin; Zhao, Chunyu; Wu, Yunchao; Chen, Qiming; Yu, Shuo; Li, Jinlong; He, Jialu; Fang, Yang; Premarathne Maha Ranvilage, Chanaka Isuranga; Huang, Xiaoping (2020). "Historical changes in seagrass beds in a rapidly urbanizing area of Guangdong Province: Implications for conservation and management". Global Ecology and Conservation. 22: e01035. doi:10.1016/j.gecco.2020.e01035. S2CID 216519471.
- ^ Jeyabaskaran, R.; Jayasankar, J.; Ambrose, T. V.; Vineetha Valsalan, K. C.; Divya, N. D.; Raji, N.; Vysakhan, P.; John, Seban; Prema, D.; Kaladharan, P.; Kripa, V. (2018). "Conservation of seagrass beds with special reference to associated species and fishery resources" (PDF). Journal of the Marine Biological Association of India. 60: 62–70. doi:10.6024/jmbai.2018.60.1.2038-10. S2CID 134131448.
- ^ Duarte, Carlos M.; Marbà, Núria; Gacia, Esperança; Fourqurean, James W.; Beggins, Jeff; Barrón, Cristina; Apostolaki, Eugenia T. (2010). "Seagrass community metabolism: Assessing the carbon sink capacity of seagrass meadows". Global Biogeochemical Cycles. 24 (4): n/a. Bibcode:2010GBioC..24.4032D. doi:10.1029/2010GB003793. S2CID 2689693.
- ^ Duarte, C. M.; Middelburg, J. J.; Caraco, N. (2005). "Major role of marine vegetation on the oceanic carbon cycle". Biogeosciences. 2 (1): 1–8. Bibcode:2005BGeo....2....1D. doi:10.5194/bg-2-1-2005.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e f Cite error: The named reference
Conte2021was invoked but never defined (see the help page).
The Holobiont Concept
[edit]Host–microbes interactions play crucial roles in biological and ecological functions, thus, organisms are better considered as a network of interactions between the host and all the associated microorganisms (bacteria, fungi and viruses), with which the host establishes transient or lasting complex relationships [40,41,42]. The host and the entirety of microorganisms living in/on its tissues represent a complex functional unit, the holobiont [33].[1]
Lynn Margulis [43,44,45] was the first to emphasize the role of the symbiosis, and in particular the endosymbiosis, as an evolutionary trajectory. Her studies are considered the starting point of this research field, despite similar ideas having already circulated years before, due to the less known German scientist Meyer-Abich [46]. Working mostly on microbial communities of corals, Zilber-Rosenberg and Rosenberg implemented this concept, considering the holobiont as an additional organismal level on which natural selection may operate [47,48], and defining the hologenome as the integration of the host genome with the gene pool of the associated microorganisms [47,49,50,51]. In this review, we will focus on the interactions between the host (seagrass) and the bacterial partners but, of course, also other microorganisms may contribute to the holobiont.[1]
The role of the host–microbe interactions, as an evolutionary driving force, is still unclear [48,50,51,52]; however, the importance of considering such interactions in the host physiology and ecology is indubitable [53,54]. In fact, the holobiont changes according to the environmental changes and maintains the host–microbes homeostasis, and its disruption may lead to pathologic conditions [55,56,57,58]. For instance, the rapid changes in the microbiota, in terms of changing community structure and composition, mutations or horizontal gene transfer, facilitate the holobiont adaptation to the continuous and unpredictable changing environmental conditions [24,52,54]. Thus, the holobiont is a theoretical and experimental framework to study the interactions between the host and its associated microbial communities in all types of ecosystems [53], from humans [59,60,61] to animals and plants [54,62,63,64,65,66,67,68,69].[1]
In terrestrial plants, the role of microbes in plants’ growth, development, nutrient uptake and defense mechanisms has been widely assessed, and important mutualistic, commensal and pathogenic interactions have been observed [54,67,68,69]. For instance, rhizobial bacteria fix nitrogen within the root nodules of their symbiotic plant partner, providing many plant species with an essential source of bioavailable nitrogen [70,71]. Among marine species, the holobiont concept has been applied to corals, sponges and seaweeds [33,63,64,65,66,72,73,74]; in many cases the variation of the associated microbiota was found as a response to environmental stress, leading to dysbiosis (microbial imbalance) [57,58,75,76,77]. Recently, the seagrass holobiont has been explored [24,25,78,79,80], with interesting but contrasting outcomes regarding the factors involved in shaping these interactions, as summarized in the next paragraphs (Section 1.2 and Section 2).[1]
- references
- ^ a b c d e Conte, Chiara; Rotini, Alice; Manfra, Loredana; d'Andrea, Marco Maria; Winters, Gidon; Migliore, Luciana (2021). "The Seagrass Holobiont: What We Know and What We Still Need to Disclose for Its Possible Use as an Ecological Indicator". Water. 13 (4): 406. doi:10.3390/w13040406.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
The Seagrass Holobiont
[edit]Microbes and seagrass establish symbiotic relationships constituting a functional unit called the holobiont that reacts as a whole to environmental changes. Recent studies have shown that the seagrass microbial associated community varies according to host species, environmental conditions and the host’s health status, suggesting that the microbial communities respond rapidly to environmental disturbances and changes. These changes, dynamics of which are still far from being clear, could represent a sensitive monitoring tool and ecological indicator to detect early stages of seagrass stress.[1]
The seagrass holobiont is made up of the plant and its associated microbial communities: seagrass harbor different and rich epiphytic microbial communities on their above- (leaves, otherwise called phyllosphere) and belowground (rhizomes and roots, otherwise called rhizosphere) plant parts [78,79,80], but also a more intimate association (endophytes) has been proposed for microbes and roots tissues [81]. Seagrass and microbes may establish symbiotic relationships [78,79,80,81]: for instance, microbes are known to: (i) enhance the nutrients and vitamins availability, which are limiting factors for seagrass growth and primary production [30,82,83,84,85,86,87,88], (ii) enhance seagrass growth, by producing hormone-like compounds [27,88] and (iii) protect seagrass roots, by detoxifying the rhizosphere [26,29,85].[1]
A remarkable difference among the microbial communities associated with the above- and belowground seagrass plant compartments has been found [24,25,31,37,39]. This is not surprising, as the two parts are positioned in very different environments in terms of light, oxygen, redox gradient and carbon availability. Furthermore, the seagrass plants themselves establish diverse chemical microenvironments: leaves release organic carbon, producing organic carbon enriched habitats [78]; roots supply their surrounding sediment with oxygen, creating aerobic microzones in the anoxic sediment and redox gradients that lead to phosphorus and iron mobilization into the rhizosphere [26,29,84]. The aboveground compartment is colonized by a large variety of generalist, aerobic organo-heterotrophic taxa, able to degrade common plants’ polymers, waste compounds and biofilm [25,78]. The belowground compartment, due to the partial presence of oxygen, harbors both anaerobic and aerobic microorganisms, such as chemolithotrophic, sulfur-oxidizing and nitrogen-fixing microorganisms [25,26,27,28,78]. Due to the radial loss of oxygen and to the release of exudates [26,89,90], the belowground compartment stimulates a selective microbial growth and in turn, receives benefits from bacterial metabolism [91,92]. Few studies have analyzed the rhizomes as a separate plant part [37,93] finding that it harbors unique microbial communities, although some microbial groups are shared with roots and sediment. As a final point, Hurtado-McCormick et al. [93] hypothesized that the local physiological activities of the seagrass may produce a further differentiation of the colonized surfaces in microhabitats. They did not find significant differences among leaves’ microhabitats (i.e., upper or lower side of the leaf), although they highlighted clear differences among rhizome/roots colonizers, probably depending on local selective environmental conditions (mentioned above).[1]
The genetic/metabolic versatility of microbes is one of the key points of the plant/microbes association [27,94,95]. Microbe versatility contributes to the plant nutrient supply, acting in the sulfur and nitrogen cycles [85,91,92,96,97]; this represents a benefit for seagrasses and allows us to infer, which are the main metabolic patterns that are taking place around the roots of seagrasses. For instance, the nifH gene (marker of nitrogenase activity) that demonstrates the presence of nitrogen-fixing bacteria, found in the microbial community associated with roots of P. oceanica, was found to belong to sulfur-oxidizing or sulphate-reducing bacteria [92], suggesting that the nitrogen-fixing bacteria may play different roles, other than being an important source of nitrogen for seagrasses [95,96,97,98,99]. Similarly, a metagenomic study on the bacteria associated with Zostera marina’s roots highlighted the presence of several genes involved in sulfur oxidation, nitrate reduction and carbon fixation [27,28].[1]
Seagrass sediments contain organic matter due to plant debris, benthic or sink dead organisms that sunk to the bottom, and to root exudates [80]. In sediments, microbial mineralization can be either aerobic, in the thin layers of the upper sediment and around the seagrass’ oxygen leaking young root tips, or anaerobic in deepest sediment layers, beyond the effect of the seagrasses’ roots [26,99,100]. The anaerobic decomposition involves sulfate-reducing bacteria and leads to the accumulation of phytotoxic H2S into the sediment, a mechanism was suggested to be responsible for historical events of wide seagrasses die-off [29,85,101]. Seagrasses tolerate low concentration of H2S [85,101,102], and they overcome its toxicity by translocating the photosynthetically produced oxygen, from the leaves to the rhizosphere supporting local spontaneous H2S reoxidation [103,104]. This allows the thriving of sulphate-oxidizing bacteria [26,28,89], found to be less abundant in low-light conditions [29].[1]
Diazotrophic bacteria, which enhance seagrass nitrogen availability [30,86,98,105,106,107], have been found in both the above- and the belowground seagrass tissues and are more abundant in vegetated sediment than in the bare one [95,107]. For instance, cyanobacteria were found to be more abundant on seagrass leaves from oligotrophic environments [35,107], and Tarquinio et al. [30] demonstrated that leaves of the temperate seagrass Posidonia sinuosa with associated microbes, accumulate more nitrogen than those devoid of microorganisms. In this laboratory study, Tarquinio et al. [30] incubated seagrass leaves with amino acids labeled with stable isotope 15N and found that the marked amino acids were assimilated by the associated microbes and by plants, but not by plants without microorganisms. This demonstrated the active role of seagrass associated microbiota both in supplying nitrogen to plants and in amino acid mineralization.[1]
Nitrogen fixing bacteria play another important role in the seagrass holobiont, being also involved in the phytotoxic sulfate-reduction, a key-role in organic matter mineralization. The seagrass release of photosynthates translocated from roots to the rhizosphere sustains epibionts associated to their tissues or in the close surrounding environment [26,29,89,107], suggesting a mutualistic relationship between the seagrass host and its diazotrophic bacteria, and the role of these microorganisms in maintaining seagrass health [30,32].[1]
Another key point in the holobiont dynamics is that seagrass might select their epiphytes and contrast pathogens, by producing antifouling and antimicrobial compounds [108,109,110]. On the other hand, even microbes produce antimicrobial compounds and may enhance seagrass defenses [27,110,111,112,113,114,115]. In fact, they produce compounds to control biofilm forming-bacteria [112,113,114,115,116,117,118] or lytic enzymes, as agarases and carrageenases, which degrade galactose-based algal polymers [119], potentially controlling the growth of microalgal biofilm [27]. For instance, members of the genus Bacillus and Virgibacillus found associated with the tropical seagrasses Thalassia hemprichii and Enhalus acoroides were shown to exert antifouling activities against biofilm-forming bacteria [117,118]. In addition, Actinobacteria, commonly found associated with seagrass [27,31,35,39,95,99,112,117], has been considered as a source of bioactive natural compounds [112]. A further important interaction in the holobiont regards the capability of microbes to break down phytotoxic compounds [26,27,32,78,79,104], contributing further to seagrass health.[1]
Thus, the microenvironments of seagrass surface could be a selective substrate for microbial growth, although this hypothesis deserves further investigation. Research in this field will surely provide new insights into plant ecology and associated microbial community dynamics, an important step towards seagrass monitoring and conservation [32,34,79].[1]
A last critical point for the holobiont comprehension is to understand the process of microbial colonization of the seagrass compartments. It must rely on the environmental microbial pool, as so far no other microbial sources have been detected [120]. This colonization process may follow different pathways: microbial communities associated with leaves generally mirror those present in the surrounding seawater column, while microbial communities associated with the rhizome/root usually strongly differs from the sediment microbial community [120].[1]
A recent study by Kohn et al. [121] focused on the variation of microbial communities on the leaves of the same Posidonia oceanica plants over time: the authors showed that a macroscopically different biofilm is found in young and older leaves, with increased diversity in older leaves, but with very similar taxonomic compositions. The influence of age on root-associated microbial biofilm was investigated but no clear trends were found [22,122]. However, the colonization time is probably an important factor shaping the seagrass holobiont, and deserves further attention.[1]
Looking at the ecosystem level, interactions among seagrasses and microorganisms can directly influence large-scale biogeochemical processes, including coastal carbon sequestration [123,124,125,126,127,128,129,130,131,132,133], the so-called blue carbon [124]. Seagrass ecosystems are significant carbon sinks, as both living plant biomass and recalcitrant dead organic matter, and the degradative activity of the sediment microbial communities control the amount of sequestered carbon [128,131]. Recent studies showed an increased mineralization rate of organic carbon in sediments as a response to eutrophication and warming seawater temperatures. In fact, both eutrophication and warming seawater temperatures may stimulate microbial metabolism and speed-up the mineralization processes, enhancing CO2 release within the water column [126,129,131,132,134]. Contrasting results were found by investigating whether the nutrient input stimulates the mineralization process in vegetated sediments [129,131]. These results suggest the co-occurrence of other factors that are able to drive the microbial response to increased nutrient availability [131]. On the contrary, rising temperatures seem to have a significant effect on the aerobic mineralization of organic carbon, but a negligible effect on both the anaerobic mineralization and the recalcitrant seagrass dead tissue mineralization, as debris of rhizomes and roots [126,131,132,133,134]. Thus, the aerobic mineralization of exposed buried carbon (as in the case of sediment resuspension by trawling) may reduce the carbon sink capacity of seagrass dead organic matter, through increased microbial abundance and speed-up of the mineralization process [133,134]. Hence, seagrass–microbes interactions may clearly affect ecosystem processes and these interactions, usually evaluated at small scales, have to be upscaled and evaluated even at a wide seascape. To this end, the study of seagrass holobiont variations in the field and/or under controlled laboratory conditions is pivotal to point out which factors shape and regulate these interactions. Such studies could contribute to seagrass monitoring and conservation efforts.[1]
- references
Holobiont before Lynn Margulis
[edit]Overview
[edit]Holobiont is a concept that has been an attractor for several ideas, and researchers have independently converged on this word to describe the integrated composite organism composed of microbial and host eukaryotic species. In evolutionary developmental biology, particularly in eco‐devo and eco‐evo‐devo, recent conceptualizations of developmental and evolutionary systems as holobionts made possible new theoretical insights and showcased heuristic fruitfulness.[1][2][3][4][5][6][7]
In 2008, Zilber‐Rosenberg and Rosenberg [8] put forth this notion to describe corals as holobionts, consisting of cnidarians plus their symbiotic algae. However, before they had proposed the term, Lynn Margulis had already used it as early as 1990.[9] Although she did not use the term frequently then or thereafter, in the last 15 years, her name has become synonymous with the concept of holobiont, and numerous papers have especially cited her 1991 chapter in the book Symbiosis as a Source of Evolutionary Innovation as the origin of the concept.[10] Since "Lynn Margulis' name is as synonymous with symbiosis as Charles Darwin's is with evolution",[11] her use of the holobiont concept and her life‐long work on symbiogenesis more generally, has become the collective starting point for a rapidly growing research field on host‐microbe interactions and hologenomes. Nevertheless, Margulis was not the first person to introduce the concept of holobiont; nor was she the first to study the development and evolution of holobionts. In this commentary we show who did, and why this matters.[7]
The holobiont concept
[edit]Today, holobiont usually refers to a close association between different individuals, usually host‐microbiota symbioses, that together form anatomical, physiological, immunological or evolutionary units.[12][13][14] In the last 10 years, the holobiont concept has rapidly become popular, not least through the associated usage of the hologenome concept.[15][16][8]
COPY from hologenome theory of evolution...
In September 1994, Richard Jefferson coined the term hologenome when he introduced the hologenome theory of evolution at a presentation at Cold Spring Harbor Laboratory.[17][18][19]
Independently coined by Jefferson during a symposium lecture in 1994,[20] the hologenome describes the entire metagenome of a holobiont. The concept has given birth to a lively and rapidly growing research community that studies how holobiont components are assembled and transmitted,[21][22][23] how this collective evolves through natural selection,[15][24][25][26] how host and microbiota metabolically interact,[27][28] and how microbiota affect host health.[29][30] But while this young research field is highly productive and growing rapidly, it has adopted an identity‐shaping origin myth that needs to be more fully explored.[7]
Even today, different concepts of super‐organismality, like "holobiont" and "metaorganism", are used in sometimes ambiguous and contradicting ways.[31][12] Moreover, the history of these terms (and their underlying ideas) is often quite complicated. For example, the German botanist and philosopher Johannes Reinke introduced the term "Konsortium" (consortium) to denote the mutual relationship of algae and fungi that establishes the super‐organismal unit of lichens.[32][33] Later, botanist and mycologist Albert Frank (1877) presented the term “Homobium” to designate a system in which partner organisms form a new, interdependent organism.[34] These are only two examples of many that could be cited, as late 19th‐century, German‐speaking biology had various theoretical discussions about symbiosis, among authors like Anton de Bary, Eduard Strasburger and Simon Schwendener.[35][36] Due to this complexity one has to be as precise as possible about what each of these concepts refers to, that is, what super‐organismal unit is being theorized. In this paper we will only focus on the history of the concept of holobiont, as it introduced an explicit evolutionary (and evo‐devo) perspective to older symbiosis discussions. The next section briefly covers the traditional narrative about the historical roots of this concept.[7]
History
[edit]Lynn Margulis and symbiogenesis
[edit]In 1905 the Russian botanist Konstantin Mereschkowski argued in his endosymbiotic theory (by drawing on earlier work on symbiosis) that eukaryotic cells evolved through symbiosis from single‐celled prokaryotes.[37] Subsequently, this and other approaches formulated until the late 1920s [38][39] were widely neglected for many decades (or at least so the standard story goes) by the then‐dominant autogenic hypothesis or direct filiation theory. It argued that mitochondrial evolution and the establishment of pro-eukaryotic cells was made possible through the slow process of invagination of the prokaryote cell membrane.[7]
From the late 1960s onwards, Margulis' evolutionary endosymbiotic theory,[40][10][41][42][10][43] drew on the earlier works of Mereschkowski, Wallin, and others,[10][41] and advanced them with microbiological evidence. While a couple of Margulis' ideas varied throughout her long career,[44] some central arguments remained unchanged.[45] These are:
(1) Symbiogenesis provides a theory of the origins of new phylogenetic forms and biological innovations, like new tissues, organs, and physiologies.[7]
(2) The fusion of genomes is the central mechanism of endosymbiosis.[7]
(3) The theory of endosymbiosis opposes the neo‐Darwinian view that evolution occurs through the accumulation and selection of mutational changes to the nuclear genome.[7]
In the early 1990s, as an extension of her theory, Margulis referred to the concept of holobiont as "the symbiotic complex" [10] or "the product, temporary or permanent, of the association between its constituent bionts",[41] but without making any reference to previous coinage or usages of the concept. In the next 15 years, the concept was at first largely neglected, but then started spreading widely. Today, the above references are consistently listed in holobiont, hologenome, and microbiome literature together with the claim that Margulis introduced the concept.[12][15][25][22][16] In introduction and background sections of articles these references function as a historical stepping stone that links Margulis' older work with more recent holobiont research. Although much of contemporary holobiont research builds on Margulis' framework, it is not the only theoretical stance available for holobiont studies. Indeed, the original framework for holobionts was introduced by the theoretical biologist Adolf Meyer‐Abich, 50 years before Margulis.[7]
Adolf Meyer‐Abich and holobiosis
[edit]Adolf Meyer‐Abich (1893–1971), also known as Adolf Meyer before 1938, was a German theoretical biologist and philosopher of science.[46] He held academic positions in Hamburg (1930–1958), as well as intermittently in Santiago de Chile, the Dominican Republic, El Salvador, and the United States (e.g., as a visiting professor at the University of Texas at Austin, 1960). Meyer‐Abich was a leading figure in early German theoretical biology, especially through his life‐long attempt to replace reductionist theories in biology through holistic approaches that interrelate evolution and development. He was also one of the founders of the journal Acta Biotheoretica.[7]
A topic he published on most extensively was his theory of holobiosis.[47][48][49][50][51] His aim was to "provide the foundation for a new evolutionistic theory of phylogenesis on the basis of the principle of holobiosis".[47] He stated:
- In a nutshell, the theory of holobiosis says that all higher and more complex organisms have developed through biontic processes, that is, parabiosis, antibiosis, symbioses, and finally holobiosis between simpler and lower forms of organisms. In other words, this means that the organs and the unions of organs have developed from originally independent organisms .[47]
Meyer‐Abich argued that to explain evolutionary change, one has to focus on describing the processes of assemblage of independent organisms, first as symbiotic partners, then holobionts (“Holobionten”), and finally systems of organs that contribute to a larger integrated whole. At the crucial evolutionary stage of holobiosis, the originally independent units are so closely interlinked that they cannot develop, grow, nor reproduce independently of one another.[7]
Meyer‐Abich used the holobiont concept slightly different than Margulis. He usually described holobionts as specific bionts within a strongly integrated whole, a holobiome (“Holobiom”) created through holobiosis, rather than as the whole itself. However, he addressed the same developmental and evolutionary phenomena as Margulis and, more important, his theory includes all three central claims later famously defended by her.[7]
(1) Holobionts, in contrast to mere symbionts, are able to realize unique traits together (like novel metabolites and reproductive bodies in lichens) that no individual organism could produce on its own. He argued that the polyphyletic diversity of holobionts allows novel forms of energy‐sharing, which guarantees fitness advantages and access to unexplored ecological niches. To support this argument, Meyer‐Abich extensively analyzed trait combinations across different phylogenetic groups, like flagellate protozoans, Burseraceae and Crustacea, to show how more “primitive” and more “differentiated” traits are always interlinked with one another in one taxon.[47][49][50][51] He argued that such trait complexes could have only originated through holobiosis. For example, he postulated that the different cell types in sponges are a result of holobiosis, that all higher plants originate from algal‐fungal holobioses (fungal components allowed the invasion of the land as rooting and absorbing organs), and that tissue differentiation and specialization in metazoans was driven through the polyphyletic diversity of holobionts.[7]
(2) Similar to Margulis, Meyer‐Abich assumed that new characters can only evolve "by causal holistic genom [sic] combinations.[51] He was convinced that genetic variation within monophyletic groups is not sufficient to wholly account for the origin of novel traits and taxa. He argued (in a somewhat cryptic style) that through holobiosis new symbionts get integrated into the plasmatic or genetic heredity apparatus of the host. This then releases new energy potentials that allow the development of more complex traits (and functions) via new mutations that could not be realized before. The assumed mechanism reminds one of what some of his contemporaries have described as the release of cryptic genetic variation.[52]
(3) Meyer‐Abich, like Margulis, believed that his theory opposed to the orthodox neo‐Darwinian framework of evolutionary theory. He argued that to understand macroevolutionary change one should not (only) try to find missing links in the fossil record. Rather, one should compare changes in developmental physiology in juvenile organisms.[47][49] This includes, especially, changes in energy potentials during embryogenesis, for instance, though the formation of novel metabolically integrated holobionts (lichens) or even stronger, morphologically integrated holobionts (sponges).[7]
Even though Meyer‐Abich anticipated Margulis' three central claims, in contrast to her, he was not able to provide much empirical data in support of his views, especially on the genetic mechanisms of holobiosis. However, he was fully aware of this, as he understood his theory of holobiosis as a fruitful "hypothesis which will guide us to new valuable research"[51][7]
Why Meyer‐Abich was forgotten
[edit]Why does today's holobiont researchers know nothing about Meyer‐Abich and thus of its own conceptual and theoretical origin? This is especially puzzling since Meyer‐Abich published extensively—around 400 pages—on his theory of holobiosis and holobionts. Nonetheless, articles and books on the history of the endosymbiotic theory or Margulis' work do not discuss Meyer‐Abich at all.[41][44][35][11][53][36] Those few historians who knew (or still know) of his work (or at least his English book from 1964) unfortunately predated the recent renaissance of the holobiont concept,[54] or did not link his ideas to Margulis' theory of symbiogenesis and holobionts.[46][55][56] To our knowledge there is just one publication in the current holobiont literature that dedicates (unfortunately merely) one sentence to Meyer‐Abich: "Holobiont is an old term in the symbiosis literature for the entity made up of symbiotic partners [49]".[57][7]
This nearly complete neglect of the conceptual and theoretical origins of holobiont research might be due to, at least, two reasons: First, after WWII, German holistic biology became suspicious. Many of its advocates, including Meyer‐Abich, had (at least at some point) endorsed National Socialism. In fact, while none of Meyer‐Abich's writings on holobionts include clear ideological statements, on several occasions he attempted to sell his work to the Nazis as a biology that fits their ideology, but ultimately failed in doing so.[58] This makes many biologists still today skeptical about reading this body of literature. Second, most of his publications on holobionts are in German. His only English‐language work The Historical‐Philosophical Background of the Modern Evolution‐Biology,[51] a series of lectures held at the University of Texas at Austin in 1960, did not have an impact on the scientific community in the United States. It is very likely that the young post‐doc Lynn Margulis (then Lynn Sagan) did not read Meyer‐Abich's book before publishing her ground‐breaking paper 3 years later,[43] nor that she ever encountered it later in her career, despite her broad knowledge about the history of the endosymbiotic theory.[7]
Conclusions
[edit]
(Not all scientists involved in these developments are mentioned)
Meyer‐Abich's forgotten theory of holobiosis not only introduced the concept of holobiont long before Lynn Margulis did. It also highly resembles Margulis' account. This is likely due to the fact that both developed their views based on the same early 20th century endosymbiotic theories, like Mereschkowski's and Wallin's.[48][49] However, Meyer‐Abich did so in a time (1940–60s) before data showed that mitochondria and bacteria shared circular DNA, 70S ribosomes, and electron transport chains. Thus, from the early endosymbiotic theories, we can postulate two lines leading to holobionts (see disgram).[7]
The first was the immediate line to the concept of holobiosis by Meyer‐Abich in the context of early endosymbiotic theories and holistic biology.[59][55][56] Meyer‐Abich's theory anticipated many of today's most central research questions about holobionts (see dotted line in Figure 3). This includes the question how their components assemble, how they interact, how holobionts evolve, and what role they might play in speciation and macroevolution.[4][60][61][8] Moreover, the very species studied by Meyer‐Abich are currently used again as model organisms to investigate the development and evolution of holobionts and other host‐microbiotes collectives: lichens,[62][27] sponges [63][64] and various plants, ranging from mosses, like Marchantia spp., to orchid species.[65][66][67][68] Remarkably, current views of holobionts, both in evo‐devo and microbiology, share with Meyer‐Abich the assumption of the phylogenetic ubiquitous nature and evolutionary centrality of holobiosis.[3][29][69][25] For him, nature was a "universal mycorrhiza".[47] As Gilbert et al. put it in 2012: "For animals, as well as plants, there have never been individuals[...] We are all lichens".[29][7]
The second line brings forth holobionts from early endosymbiotic theory plus the newer knowledge of DNA in cell organelles (see Figure 3). While she did not integrate Meyer‐Abich's earlier work, Margulis was able to synthesize evidence from numerous sources and integrate them together with new data to convince people that hers was an accurate way of looking at the natural world.[7]
In this paper, we focused on the history of the holobiont concept in 20th‐century biology. The term “holobiont” was independently conceived (at least two times), first by Adolf Meyer‐Abich in the 1940s and later by Margulis in the 1990s. We showed that Meyer‐Abich's view of holobiosis anticipated several key ideas of Margulis, and that both share similar theoretical underpinnings due to earlier symbiosis research studied and integrated by both authors. In this way, Meyer‐Abich should be recognized as the first scholar who introduced the holobiont concept in biology. The connections of Meyer‐Abich's conceptual framework to previous 19th‐century concepts, such as Johannes Reinke's Konsortium or Albert Frank's Homobium, deserve a separate and detailed historical treatment. The same applies to the problematic reception of the Meyer‐Abich theory after the 1960s.[7]
As the holobiont has become a central guiding concept for today's microbiology and microbiome studies in evolutionary developmental biology for organizing research, labeling conferences, and publishing papers, this interdisciplinary research community should become aware of one of its central and so‐far neglected conceptual and theoretical founders: Adolf Meyer‐Abich. The endosymbiosis “story” has been used as a tale of scientific surprise, heroism, conservatism, misogyny, and the triumph of molecular phylogenetics.[57][35] But it is more than this. It is a story about what counts as evidence at any particular time and place, and who gets to channel data into particular narratives. Contemporary science suffers from “lather, rinse, repeat”‐like strategies concerning coinage of concepts and acknowledging “true” discoverers.[70] Such errors in referencing (especially in background and introduction sections of articles) can massively shape and, in fact, distort the historical identity of young research fields. However, in holobiont research, it is not too late to correct this trend.[7]
- ^ Bosch, Thomas C. G.; Miller, David J. (8 March 2016). The Holobiont Imperative: Perspectives from Early Emerging Animals. ISBN 9783709118962.
- ^ Carrier, Tyler J.; Reitzel, Adam M. (2017). "The Hologenome Across Environments and the Implications of a Host-Associated Microbial Repertoire". Frontiers in Microbiology. 8: 802. doi:10.3389/fmicb.2017.00802. PMC 5425589. PMID 28553264.
- ^ a b Gilbert, Scott F. (2019). "Evolutionary transitions revisited: Holobiont evo‐devo". Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 332 (8): 307–314. doi:10.1002/jez.b.22903. PMID 31565856. S2CID 203608526.
- ^ a b Collens, Adena; Kelley, Emma; Katz, Laura A. (2019). "The concept of the hologenome, an epigenetic phenomenon, challenges aspects of the modern evolutionary synthesis". Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 332 (8): 349–355. doi:10.1002/jez.b.22915. PMC 6904923. PMID 31709760.
- ^ Gilbert, Scott F.; Bosch, Thomas C. G.; Ledón-Rettig, Cristina (2015). "Eco-Evo-Devo: Developmental symbiosis and developmental plasticity as evolutionary agents". Nature Reviews Genetics. 16 (10): 611–622. doi:10.1038/nrg3982. PMID 26370902. S2CID 205486234.
- ^ Schwab, Daniel B.; Casasa, Sofia; Moczek, Armin P. (2019). "On the Reciprocally Causal and Constructive Nature of Developmental Plasticity and Robustness". Frontiers in Genetics. 9: 735. doi:10.3389/fgene.2018.00735. PMC 6335315. PMID 30687394.
- ^ a b c d e f g h i j k l m n o p q r s t u v w Baedke, Jan; Fábregas‐Tejeda, Alejandro; Nieves Delgado, Abigail (2020). "The holobiont concept before Margulis". Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 334 (3): 149–155. doi:10.1002/jez.b.22931. PMID 32039567. S2CID 211070611.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c Zilber-Rosenberg, Ilana; Rosenberg, Eugene (2008). "Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution". FEMS Microbiology Reviews. 32 (5): 723–735. doi:10.1111/j.1574-6976.2008.00123.x. PMID 18549407.
- ^ Cowley, Allen W. (2004). "The elusive field of systems biology". Physiological Genomics. 16 (3): 285–286. doi:10.1152/physiolgenomics.00007.2004. PMID 14966250.
- ^ a b c d e Margulis, Lynn; Fester, René (1991). Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. ISBN 9780262132695.
{{cite book}}: Missing|author1=(help) - ^ a b Sapp, J. (2012). "Too fantastic for polite society: A brief history of symbiosis theory". In: Sagan, Dorion (2012). Lynn Margulis: The Life and Legacy of a Scientific Rebel. pp. 54–67. ISBN 9781603584463.
- ^ a b c Bordenstein, Seth R.; Theis, Kevin R. (2015). "Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes". PLOS Biology. 13 (8): e1002226. doi:10.1371/journal.pbio.1002226. PMC 4540581. PMID 26284777.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Gilbert, Scott F.; Sapp, Jan; Tauber, Alfred I. (2012). "A Symbiotic View of Life: We Have Never Been Individuals". The Quarterly Review of Biology. 87 (4): 325–341. doi:10.1086/668166. PMID 23397797. S2CID 14279096.
- ^ Simon, Jean-Christophe; Marchesi, Julian R.; Mougel, Christophe; Selosse, Marc-André (2019). "Host-microbiota interactions: From holobiont theory to analysis". Microbiome. 7 (1): 5. doi:10.1186/s40168-019-0619-4. PMC 6330386. PMID 30635058. S2CID 58012049.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c Morris, J. Jeffrey (2018). "What is the hologenome concept of evolution?". F1000Research. 7: 1664. doi:10.12688/f1000research.14385.1. PMC 6198262. PMID 30410727.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Theis, Kevin R.; Dheilly, Nolwenn M.; Klassen, Jonathan L.; Brucker, Robert M.; Baines, John F.; Bosch, Thomas C. G.; Cryan, John F.; Gilbert, Scott F.; Goodnight, Charles J.; Lloyd, Elisabeth A.; Sapp, Jan; Vandenkoornhuyse, Philippe; Zilber-Rosenberg, Ilana; Rosenberg, Eugene; Bordenstein, Seth R. (2016). "Getting the Hologenome Concept Right: An Eco-Evolutionary Framework for Hosts and Their Microbiomes". mSystems. 1 (2). doi:10.1128/mSystems.00028-16. PMC 5069740. PMID 27822520.
- ^ Jefferson, Richard (2019-04-04). "Agriculture and the Third World". figshare. doi:10.6084/m9.figshare.7945781. Retrieved 2019-04-04.
- ^ Number 6 in a series of 7 VHS recordings, A Decade of PCR: Celebrating 10 Years of Amplification, Cold Spring Harbor Laboratory Press, 1994. ISBN 0-87969-473-4. http://www.cshlpress.com/default.tpl?..
- ^ Part 4: The Hologenome - YouTube Agriculture, Environment and the Developing World: A Future of PCR. Part 4: The Hologenome Plenary lecture by Richard Jefferson, CEO Cambia, Cold Spring Harbor, September, 1994.
- ^ Jefferson, R. (1994). "The hologenome: Agriculture, environment and the developing world: A future of PCR". New York.
- ^ Gilbert, Scott F. (2014). "A holobiont birth narrative: The epigenetic transmission of the human microbiome". Frontiers in Genetics. 5: 282. doi:10.3389/fgene.2014.00282. PMC 4137224. PMID 25191338.
- ^ a b Sitaraman, Ramakrishnan (2018). "Prokaryotic horizontal gene transfer within the human holobiont: Ecological-evolutionary inferences, implications and possibilities". Microbiome. 6 (1): 163. doi:10.1186/s40168-018-0551-z. PMC 6142633. PMID 30223892. S2CID 52291350.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Vannier, Nathan; Bittebiere, Anne-Kristel; Vandenkoornhuyse, Philippe; Mony, Cendrine (2016). "AM fungi patchiness and the clonal growth of Glechoma hederacea in heterogeneous environments". Scientific Reports. 6: 37852. Bibcode:2016NatSR...637852V. doi:10.1038/srep37852. PMC 5122940. PMID 27886270.
- ^ Huitzil, Saúl; Sandoval-Motta, Santiago; Frank, Alejandro; Aldana, Maximino (2018). "Modeling the Role of the Microbiome in Evolution". Frontiers in Physiology. 9: 1836. doi:10.3389/fphys.2018.01836. PMC 6307544. PMID 30618841.
- ^ a b c Rosenberg, Eugene; Zilber-Rosenberg, Ilana (2018). "The hologenome concept of evolution after 10 years". Microbiome. 6 (1): 78. doi:10.1186/s40168-018-0457-9. PMC 5922317. PMID 29695294. S2CID 13680513.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Roughgarden, Joan; Gilbert, Scott F.; Rosenberg, Eugene; Zilber-Rosenberg, Ilana; Lloyd, Elisabeth A. (2018). "Holobionts as Units of Selection and a Model of Their Population Dynamics and Evolution". Biological Theory. 13: 44–65. doi:10.1007/s13752-017-0287-1. S2CID 90306574.
- ^ Postler, Thomas Siegmund; Ghosh, Sankar (2017). "Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System". Cell Metabolism. 26 (1): 110–130. doi:10.1016/j.cmet.2017.05.008. PMC 5535818. PMID 28625867.
- ^ a b c Gilbert, Scott F.; Sapp, Jan; Tauber, Alfred I. (2012). "A Symbiotic View of Life: We Have Never Been Individuals". The Quarterly Review of Biology. 87 (4): 325–341. doi:10.1086/668166. PMID 23397797. S2CID 14279096.
- ^ Van De Guchte, Maarten; Blottière, Hervé M.; Doré, Joël (2018). "Humans as holobionts: Implications for prevention and therapy". Microbiome. 6 (1): 81. doi:10.1186/s40168-018-0466-8. PMC 5928587. PMID 29716650. S2CID 13660084.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Bang, Corinna; Dagan, Tal; Deines, Peter; Dubilier, Nicole; Duschl, Wolfgang J.; Fraune, Sebastian; Hentschel, Ute; Hirt, Heribert; Hülter, Nils; Lachnit, Tim; Picazo, Devani; Pita, Lucia; Pogoreutz, Claudia; Rädecker, Nils; Saad, Maged M.; Schmitz, Ruth A.; Schulenburg, Hinrich; Voolstra, Christian R.; Weiland-Bräuer, Nancy; Ziegler, Maren; Bosch, Thomas C.G. (2018). "Metaorganisms in extreme environments: Do microbes play a role in organismal adaptation?". Zoology. 127: 1–19. doi:10.1016/j.zool.2018.02.004. PMID 29599012.
- ^ Reinke, J. (1872) "Ueber die anatomischen Verhältnisse einiger Arten von Gunnera L. Nachrichten von der Königl". Gesellschaft der Wissenschaften und der Georg-Augusts-Universität zu Göttingen, 1872, pp.100–108.
- ^ Kull, Kalevi (2010). "Ecosystems are Made of Semiosic Bonds: Consortia, Umwelten, Biophony and Ecological Codes". Biosemiotics. 3 (3): 347–357. doi:10.1007/s12304-010-9081-1. S2CID 28361059.
- ^ Frank, A. B. (1877). "Ueber die biologischen Verhältnisse des Thallus einiger Krustenflechten". [On the biological characteristics of the thallus of some crustose lichens]. Beiträge zur Biologie der Pflanzen, 2: 123–200.
- ^ a b c Sapp, Jan (1994). Evolution by Association: A History of Symbiosis. ISBN 9780195088212.
- ^ a b Suárez, Javier (2018). "'The importance of symbiosis in philosophy of biology: An analysis of the current debate on biological individuality and its historical roots'". Symbiosis. 76 (2): 77–96. doi:10.1007/s13199-018-0556-1. S2CID 13786151.
- ^ Mereschkowski, K. (1905). "Über Natur und Ursprung der Chromatophoren im Pflanzenreich" [About the nature and origin of chromatophores in the plant kingdom]. Biologisches Centralblatt, 25: 593–604.
- ^ Mereschkowski, K. (1905) "Über Natur und Ursprung der Chromatophoren im Pflanzenreich" [About the nature and origin of chromatophores in the plant kingdom]. Biologisches Centralblatt, 25: 593–604.
- ^ Wallin, I. E. (1927) "Symbioticism and the origin of species". Baltimore: Williams & Wilkins Co.
- ^ Margulis, Lynn (1970). Origin of Eukaryotic Cells: Evidence and Research Implications for a Theory of the Origin and Evolution of Microbial, Plant, and Animal Cells on the Precambrian Earth. ISBN 9780300013535.
- ^ a b c d Margulis, L. (1993) "Symbiosis in cell evolution: microbial communities in the Archean and Proterozoic eons". WH Freeman, New York.
- ^ Witzany, Günther (2006). "Serial Endosymbiotic Theory (Set): The Biosemiotic Update". Acta Biotheoretica. 54 (2): 103–117. doi:10.1007/s10441-006-7831-x. PMID 16988903. S2CID 18026766.
- ^ a b Sagan, Lynn (1967). "On the origin of mitosing cells". Journal of Theoretical Biology. 14 (3): 225–IN6. doi:10.1016/0022-5193(67)90079-3. PMID 11541392.
- ^ a b Martin, William F. (2017). "Physiology, anaerobes, and the origin of mitosing cells 50 years on". Journal of Theoretical Biology. 434: 2–10. doi:10.1016/j.jtbi.2017.01.004. PMID 28087421.
- ^ o'Malley, Maureen A. (2017). "From endosymbiosis to holobionts: Evaluating a conceptual legacy". Journal of Theoretical Biology. 434: 34–41. doi:10.1016/j.jtbi.2017.03.008. PMID 28302492.
- ^ a b Amidon, Kevin S. (2008). "Adolf Meyer-Abich, Holism, and the Negotiation of Theoretical Biology". Biological Theory. 3 (4): 357–370. doi:10.1162/biot.2008.3.4.357. S2CID 43525676.
- ^ a b c d e f Meyer‐Abich, A. (1943). Beiträge zur Theorie der Evolution der Organismen. I. Das typologische Grundgesetz und seine Folgerungen für Phylogenie und Entwicklungsphysiologie [Contributions to the evolutionary theory of organisms: I. The basic typological law and its implications for phylogeny and developmental physiology]. Acta Biotheoretica, 7, 1–80.
- ^ a b Meyer‐Abich, A. (1948). Naturphilosophie auf neuen Wegen [Natural philosophy on new paths]. Stuttgart: Hippokrates‐Verlag Marquardt.
- ^ a b c d e Meyer‐Abich, A. (1950). Beiträge zur Theorie der Evolution der Organismen. II. Typensynthese durch Holobiose [Contributions to the evolutionary theory of organisms: II. Synthesis of types through holobiosis]. (Bibliotheca Biotheoretica 5). Leiden: E.J. Brill.
- ^ a b Meyer‐Abich, A. (1963). Geistesgeschichtliche Grundlagen der Biologie [Foundation of biology in the history of ideas]. Stuttgart: Gustav Fischer.
- ^ a b c d e Meyer‐Abich, A. (1964). The historico‐philosophical background of the modern evolution‐biology. (Bibliotheca Biotheoretica 10). Leiden: E.J. Brill.
- ^ Waddington, C. H. (1953). "Genetic Assimilation of an Acquired Character". Evolution. 7 (2): 118–126. doi:10.2307/2405747. JSTOR 2405747.
- ^ Simon, Jean-Christophe; Marchesi, Julian R.; Mougel, Christophe; Selosse, Marc-André (2019). "Host-microbiota interactions: From holobiont theory to analysis". Microbiome. 7 (1): 5. doi:10.1186/s40168-019-0619-4. PMC 6330386. PMID 30635058.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ . doi:https://books.google.com/books?id=MPRAYaI8zHkC&dq=Evolutionary theory: The unfinished synthesis&source=gbs_book_other_versions (inactive 2022-06-04).
{{cite journal}}: Check|doi=value (help); Cite journal requires|journal=(help); External link in|doi=|title=(help)CS1 maint: DOI inactive as of June 2022 (link) - ^ a b Peterson, Erik L. (10 February 2017). The Life Organic: The Theoretical Biology Club and the Roots of Epigenetics. ISBN 9780822981985.
- ^ a b Rieppel, Olivier (6 July 2016). Phylogenetic Systematics: Haeckel to Hennig. ISBN 9781498754897.
- ^ a b Queller, David C.; Strassmann, Joan E. (2016). "Problems of multi-species organisms: Endosymbionts to holobionts". Biology & Philosophy. 31 (6): 855–873. doi:10.1007/s10539-016-9547-x. S2CID 89188534.
- ^ Dahn, Ryan (2019). "Big Science, Nazified? Pascual Jordan, Adolf Meyer-Abich, and the Abortive Scientific Journal Physis". Isis. 110: 68–90. doi:10.1086/701352. hdl:21.11116/0000-0003-D105-B. S2CID 151049364.
- ^ Baedke, Jan (2019). "O Organism, Where Art Thou? Old and New Challenges for Organism-Centered Biology". Journal of the History of Biology. 52 (2): 293–324. doi:10.1007/s10739-018-9549-4. PMID 30465299. S2CID 53713339.
- ^ Parker, Erik S.; Dury, Guillaume J.; Moczek, Armin P. (2019). "Transgenerational developmental effects of species-specific, maternally transmitted microbiota in Onthophagusdung beetles". Ecological Entomology. 44 (2): 274–282. doi:10.1111/een.12703. S2CID 92271506.
- ^ Sevellec, Maelle; Derome, Nicolas; Bernatchez, Louis (2018). "Holobionts and ecological speciation: The intestinal microbiota of lake whitefish species pairs". Microbiome. 6 (1): 47. doi:10.1186/s40168-018-0427-2. PMC 5853090. PMID 29540239.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Aschenbrenner, Ines A.; Cernava, Tomislav; Berg, Gabriele; Grube, Martin (2016). "Understanding Microbial Multi-Species Symbioses". Frontiers in Microbiology. 7: 180. doi:10.3389/fmicb.2016.00180. PMC 4757690. PMID 26925047.
- ^ Pita, Lucia; Fraune, Sebastian; Hentschel, Ute (2016). "Emerging Sponge Models of Animal-Microbe Symbioses". Frontiers in Microbiology. 7: 2102. doi:10.3389/fmicb.2016.02102. PMC 5179597. PMID 28066403.
- ^ Webster, Nicole S.; Thomas, Torsten (2016). "The Sponge Hologenome". mBio. 7 (2): e00135-16. doi:10.1128/mBio.00135-16. PMC 4850255. PMID 27103626.
- ^ Alcaraz, Luis D.; Peimbert, Mariana; Barajas, Hugo R.; Dorantes-Acosta, Ana E.; Bowman, John L.; Arteaga-Vázquez, Mario A. (2018). "Marchantia liverworts as a proxy to plants' basal microbiomes". Scientific Reports. 8 (1): 12712. doi:10.1038/s41598-018-31168-0. PMC 6107579. PMID 30140076.
- ^ Hassani, M. Amine; Durán, Paloma; Hacquard, Stéphane (2018). "Microbial interactions within the plant holobiont". Microbiome. 6 (1): 58. doi:10.1186/s40168-018-0445-0. PMC 5870681. PMID 29587885.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Jacquemyn, Hans; Waud, Michael; Merckx, Vincent S. F. T.; Lievens, Bart; Brys, Rein (2015). "Mycorrhizal diversity, seed germination and long-term changes in population size across nine populations of the terrestrial orchid Neottia ovata". Molecular Ecology. 24 (13): 3269–3280. doi:10.1111/mec.13236. PMID 25963669. S2CID 9145575.
- ^ Vandenkoornhuyse, Philippe; Quaiser, Achim; Duhamel, Marie; Le Van, Amandine; Dufresne, Alexis (2015). "The importance of the microbiome of the plant holobiont". New Phytologist. 206 (4): 1196–1206. doi:10.1111/nph.13312. PMID 25655016.
- ^ McFall-Ngai, Margaret; Hadfield, Michael G.; Bosch, Thomas C. G.; Carey, Hannah V.; Domazet-Lošo, Tomislav; Douglas, Angela E.; Dubilier, Nicole; Eberl, Gerard; Fukami, Tadashi; Gilbert, Scott F.; Hentschel, Ute; King, Nicole; Kjelleberg, Staffan; Knoll, Andrew H.; Kremer, Natacha; Mazmanian, Sarkis K.; Metcalf, Jessica L.; Nealson, Kenneth; Pierce, Naomi E.; Rawls, John F.; Reid, Ann; Ruby, Edward G.; Rumpho, Mary; Sanders, Jon G.; Tautz, Diethard; Wernegreen, Jennifer J. (2013). "Animals in a bacterial world, a new imperative for the life sciences". Proceedings of the National Academy of Sciences. 110 (9): 3229–3236. doi:10.1073/pnas.1218525110. PMC 3587249. PMID 23391737.
- ^ Lyons, Sherrie L. (20 July 2020). From Cells to Organisms: Re-envisioning Cell Theory. ISBN 9781442635098.
Omics
[edit]- Main Biological Aerosols, Specificities, Abundance, and Diversity
- viromics: which virus are present?
- metagenomics: which microorganisms are present?
- metatranscriptomics: which microorganisms are active?
- See: Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome
Insectivorous bats
[edit]


- ^ a b c Haarsma, Anne‐Jifke; Siepel, Henk; Gravendeel, Barbara (2016). "Added value of metabarcoding combined with microscopy for evolutionary studies of mammals". Zoologica Scripta. 45: 37–49. doi:10.1111/zsc.12214. S2CID 89048681.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Metabarcoding
[edit]DNA barcoding uses short genetic sequences to identify individual taxa. By contrast, DNA metabarcoding supports simultaneous identification of entire assemblages via high-throughput sequencing.[1][2] Using metabarcoding for ecosystem monitoring provides an opportunity to identify organisms in bulk samples at a high taxonomic resolution consistently and accurately (Biomonitoring 2.0[3]).[4]
A key issue is the distinction between bulk-community sampling and environmental DNA (eDNA). eDNA samples focus on a signal derived predominantly from traces of intracellular and extracellular DNA without attempting to isolate organisms (e.g., from water or soil;[5][6]), whereas bulk-community samples include eDNA, but target the collection of whole organisms. eDNA can be effective in detecting biological signal from the environment, but the significant spatial and temporal uncertainty of that signal clouds its application in observational studies. In addition, the ease with which trace amounts of DNA can be transported makes cross-contamination a critical issue for eDNA studies (i.e., the addition of false-positives,[7] whereas the high concentrations of template material in bulk samples mean this is less of a concern.[8] As a result, our examples of metabarcoding below focus entirely on observations derived from unsorted bulk-community samples that are otherwise identical to traditional monitoring surveys.[9]
"After a decade of intensive use, it is easy to summarize the main advantages of metabarcoding: it allows for time- and cost-effective analysis of a large number of samples with high sensitivity and taxonomic resolution. This approach aims at providing an inventory of the lineages present in a sample and their relative abundances (or activities, if targeting RNA rather than DNA). However, like all methods, metabarcoding is prone to technical and biological limitations (reviewed by Taberlet et al. 2018; Santoferrara 2019)."[10]
" Metabarcoding is the community based counterpart of DNA barcoding, which instead focuses on the identification of individual taxa (ideally at the species level) using a standardized genetic marker (Hebert et al. 2003)."[10]
"although problems still exist for inference of meaningful taxonomic and ecological knowledge based on short DNA sequences"[10]
"Ten years of high-throughput protistan metabarcoding"[10]
Biological interactions and networks
[edit]Bryozoa
[edit]- Bryozoan - moss animals
- Bryozoan limestone
- Oamaru limestone
- Moeraki Boulders
Seaweed
[edit]

-
coralline algae
-
red seaweed
-
red algae
-
squid larva
-
fish eggs
Evolution of the carbon cycle
[edit]
- ^ Taberlet, Pierre; Coissac, Eric; Hajibabaei, Mehrdad; Rieseberg, Loren H. (2012). "Environmental DNA". Molecular Ecology. 21 (8): 1789–1793. doi:10.1111/j.1365-294X.2012.05542.x. PMID 22486819. S2CID 3961830.
- ^ Yu, Douglas W.; Ji, Yinqiu; Emerson, Brent C.; Wang, Xiaoyang; Ye, Chengxi; Yang, Chunyan; Ding, Zhaoli (2012). "Biodiversity soup: Metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring". Methods in Ecology and Evolution. 3 (4): 613–623. doi:10.1111/j.2041-210X.2012.00198.x. S2CID 83095866.
- ^ Baird, Donald J.; Hajibabaei, Mehrdad (2012). "Biomonitoring 2.0: A new paradigm in ecosystem assessment made possible by next-generation DNA sequencing". Molecular Ecology. 21 (8): 2039–2044. doi:10.1111/j.1365-294X.2012.05519.x. PMID 22590728. S2CID 3889647.
- ^ Bush, Alex; Monk, Wendy A.; Compson, Zacchaeus G.; Peters, Daniel L.; Porter, Teresita M.; Shokralla, Shadi; Wright, Michael T. G.; Hajibabaei, Mehrdad; Baird, Donald J. (2020). "DNA metabarcoding reveals metacommunity dynamics in a threatened boreal wetland wilderness". Proceedings of the National Academy of Sciences. 117 (15): 8539–8545. doi:10.1073/pnas.1918741117. PMC 7165428. PMID 32217735.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Deiner, Kristy; Bik, Holly M.; Mächler, Elvira; Seymour, Mathew; Lacoursière‐Roussel, Anaïs; Altermatt, Florian; Creer, Simon; Bista, Iliana; Lodge, David M.; Vere, Natasha; Pfrender, Michael E.; Bernatchez, Louis (2017). "Environmental DNA metabarcoding: Transforming how we survey animal and plant communities". Molecular Ecology. 26 (21): 5872–5895. doi:10.1111/mec.14350. PMID 28921802. S2CID 8001074.
- ^ Cristescu, Melania E.; Hebert, Paul D.N. (2018). "Uses and Misuses of Environmental DNA in Biodiversity Science and Conservation". Annual Review of Ecology, Evolution, and Systematics. 49: 209–230. doi:10.1146/annurev-ecolsys-110617-062306. S2CID 84179772.
- ^ Ficetola, Gentile F.; Pansu, Johan; Bonin, Aurélie; Coissac, Eric; Giguet-Covex, Charline; De Barba, Marta; Gielly, Ludovic; Lopes, Carla M.; Boyer, Frédéric; Pompanon, François; Rayé, Gilles; Taberlet, Pierre (2015). "Replication levels, false presences and the estimation of the presence/Absence from eDNA metabarcoding data". Molecular Ecology Resources. 15 (3): 543–556. doi:10.1111/1755-0998.12338. PMID 25327646. S2CID 24432585.
- ^ Majaneva, Markus; Diserud, Ola H.; Eagle, Shannon H.C.; Hajibabaei, Mehrdad; Ekrem, Torbjørn (2018). "Choice of DNA extraction method affects DNA metabarcoding of unsorted invertebrate bulk samples". Metabarcoding and Metagenomics. 2. doi:10.3897/mbmg.2.26664. S2CID 55137011.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Bush, Alex; Compson, Zacchaeus G.; Monk, Wendy A.; Porter, Teresita M.; Steeves, Royce; Emilson, Erik; Gagne, Nellie; Hajibabaei, Mehrdad; Roy, Mélanie; Baird, Donald J. (2019). "Studying Ecosystems with DNA Metabarcoding: Lessons from Biomonitoring of Aquatic Macroinvertebrates". Frontiers in Ecology and Evolution. 7. doi:10.3389/fevo.2019.00434. S2CID 207958228.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c d Santoferrara, Luciana; Burki, Fabien; Filker, Sabine; Logares, Ramiro; Dunthorn, Micah; McManus, George B. (2020). "Perspectives from Ten Years of Protist Studies by High‐Throughput Metabarcoding". Journal of Eukaryotic Microbiology. 67 (5): 612–622. doi:10.1111/jeu.12813. hdl:10261/223228. PMID 32498124. S2CID 219331807.
- ^ Küpper, Frithjof C.; Kamenos, Nicholas A. (2018). "The future of marine biodiversity and marine ecosystem functioning in UK coastal and territorial waters (Including UK Overseas Territories) – with an emphasis on marine macrophyte communities". Botanica Marina. 61 (6): 521–535. doi:10.1515/bot-2018-0076. S2CID 91666150.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Knoll, Andrew H.; Nowak, Martin A. (2017). "The timetable of evolution". Science Advances. 3 (5): e1603076. Bibcode:2017SciA....3E3076K. doi:10.1126/sciadv.1603076. PMC 5435417. PMID 28560344.
- ^ Catling, David C.; Kasting, James F. (13 April 2017). Atmospheric Evolution on Inhabited and Lifeless Worlds. ISBN 9780521844123.
- ^ Catling, David C.; Zahnle, Kevin J. (2020). "The Archean atmosphere". Science Advances. 6 (9): eaax1420. Bibcode:2020SciA....6.1420C. doi:10.1126/sciadv.aax1420. PMC 7043912. PMID 32133393.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Carbon cycle – Role of microorganisms
[edit]Microorganisms have key roles in carbon and nutrient cycling, animal (including human) and plant health, agriculture and the global food web. Microorganisms live in all environments on Earth that are occupied by macroscopic organisms, and they are the sole life forms in other environments, such as the deep subsurface and ‘extreme’ environments. Microorganisms date back to the origin of life on Earth at least 3.8 billion years ago, and they will likely exist well beyond any future extinction events... Unless we appreciate the importance of microbial processes, we fundamentally limit our understanding of Earth’s biosphere and response to climate change and thus jeopardize efforts to create an environmentally sustainable future.[1][2]
A microorganism is any microscopic organism or virus not visible to the naked eye (smaller than 50 μm) that can exist in a unicellular, multicellular (for example, differentiating species), aggregate (for example, biofilm) or viral form. Examples are microscopic bacteria, archaea, eukaryotes and viruses, as well as macroscopic unicellular eukaryotes, such as larger marine phytoplankton, and wood-decomposing fungi.[1]
Marine biome
[edit]Marine biomes cover ~70% of Earth’s surface and range from coastal estuaries, mangroves and coral reefs to the open oceans (Fig. 1). Phototrophic microorganisms use the sun’s energy in the top 200 m of the water column, whereas marine life in deeper zones uses organic and inorganic chemicals for energy10. In addition to sunlight, the availability of other energy forms and water temperature (ranging from approximately −2 °C in ice-covered seas to more than 100 °C in hydrothermal vents) influence the composition of marine communities11. Rising temperatures not only affect biological processes but also reduce water density and thereby stratification and circulation, which affect organismal dispersal and nutrient transport. Precipitation, salinity and winds also affect stratification, mixing and circulation. Nutrient inputs from air, river and estuarine flows also affect microbial community composition and function, and climate change affects all these physical factors.[1]
The overall relevance of microorganisms to ocean ecosystems can be appreciated from their number and biomass in the water column and subsurface: the total number of cells is more than 1029,[3][4][5][6][7][8] and the Census of Marine Life estimates that 90% of marine biomass is microbial. Beyond their sheer numbers, marine microorganisms fulfil key ecosystem functions. By fixing carbon and nitrogen, and remineralizing organic matter, marine microorganisms form the basis of ocean food webs and thus global carbon and nutrient cycles.[5] The sinking, deposition and burial of fixed carbon in particulate organic matter to marine sediments is a key, long-term mechanism for sequestering CO2 from the atmosphere. Therefore, the balance between regeneration of CO2 and nutrients via remineralization versus burial in the seabed determines the effect on climate change.[1]
In addition to getting warmer (from increased atmospheric CO2 concentrations enhancing the greenhouse effect), oceans have acidified by ~0.1 pH units since preindustrial times, with further reductions of 0.3–0.4 units predicted by the end of the century.[9][10][11] Given the unprecedented rate of pH change[11][12][13] there is a need to rapidly learn how marine life will respond.[14] The impact of elevated greenhouse gas concentrations on ocean temperature, acidification, stratification, mixing, thermohaline circulation, nutrient supply, irradiation and extreme weather events affects the marine microbiota in ways that have substantial environmental consequences, including major shifts in productivity, marine food webs, carbon export and burial in the seabed.[11][15][16][17][18][19][1]
- Microorganisms affect climate change
Marine phytoplankton perform half of the global photosynthetic CO2 fixation (net global primary production of ~50 petagram C per year) and half of the oxygen production despite amounting to only ~1% of global plant biomass.[20] In comparison with terrestrial plants, marine phytoplankton are distributed over a larger surface area, are exposed to less seasonal variation and have markedly faster turnover rates than trees (days versus decades).[20] Therefore, phytoplankton respond rapidly on a global scale to climate variations. These characteristics are important when one is evaluating the contributions of phytoplankton to carbon fixation and forecasting how this production may change in response to perturbations. Predicting the effects of climate change on primary productivity is complicated by phytoplankton bloom cycles that are affected by both bottom-up control (for example, availability of essential nutrients and vertical mixing) and top-down control (for example, grazing and viruses).[17][20][21][22][23][24] Increases in solar radiation, temperature and freshwater inputs to surface waters strengthen ocean stratification and consequently reduce transport of nutrients from deep water to surface waters, which reduces primary productivity.[17][24][25] Conversely, rising CO2 levels can increase phytoplankton primary production, but only when nutrients are not limiting.[26][27][28][1]
Some studies indicate that overall global oceanic phytoplankton density has decreased in the past century,[29] but these conclusions have been questioned because of the limited availability of long-term phytoplankton data, methodological differences in data generation and the large annual and decadal variability in phytoplankton production.[30][31][32][33] Moreover, other studies suggest a global increase in oceanic phytoplankton production [34] and changes in specific regions or specific phytoplankton groups.[35][36] The global sea ice (Sea Ice Index) is declining, leading to higher light penetration and potentially more primary production;[37] however, there are conflicting predictions for the effects of variable mixing patterns and changes in nutrient supply and for productivity trends in polar zones.[24] This highlights the need to collect long-term data on phytoplankton production and microbial community composition. Long-term data are needed to reliably predict how microbial functions and feedback mechanisms will respond to climate change, yet only very few such datasets exist (for example, the Hawaii Ocean Time-series and the Bermuda Atlantic Time-series Study).[38][39][40][40] In this context, the Global Ocean Sampling Expedition,[41] transects of the Southern Ocean,[42][43] and the Tara Oceans Consortium [44][45][46][47][48][49][50] provide metagenome data that are a valuable baseline of marine microorganisms.[1]
Diatoms perform 25–45% of total primary production in the oceans,[51][52][53] owing to their prevalence in open-ocean regions when total phytoplankton biomass is maximal.[54] Diatoms have relatively high sinking speeds compared with other phytoplankton groups, and they account for ~40% of particulate carbon export to depth.[53][55] Physically driven seasonal enrichments in surface nutrients favour diatom blooms. Anthropogenic climate change will directly affect these seasonal cycles, changing the timing of blooms and diminishing their biomass, which will reduce primary production and CO2 uptake.[56] Remote sensing data suggest a global decline of diatoms between 1998 and 2012, particularly in the North Pacific, which is associated with shallowing of the surface mixed layer and lower nutrient concentrations.[36][1]
In addition to the contribution of marine phytoplankton to CO2 sequestration,[20][57][58][59] chemolithoautotrophic archaea and bacteria fix CO2 under dark conditions in deep ocean waters [60] and at the surface during polar winter.[61] Marine bacteria and archaea also contribute substantially to surface ocean respiration and cycling of many elements.[10] Seafloor methanogens and methanotrophs are important producers and consumers of CH4, but their influence on the atmospheric flux of this greenhouse gas is uncertain.[62] Marine viruses, bacteriovorous bacteria and eukaryotic grazers are also important components of microbial food webs; for example, marine viruses influence how effectively carbon is sequestered and deposited into the deep ocean.[48] Climate change affects predator–prey interactions, including virus–host interactions, and thereby global biogeochemical cycles.[63][1]
Oxygen minimum zones (OMZs) have expanded in the past 50 years as a result of ocean warming, which reduces oxygen solubility.[64][65][66] OMZs are global sinks for reactive nitrogen, and microbial production of N2 and N2O accounts for ~25–50% of nitrogen loss from the ocean to the atmosphere. Furthermore, OMZs are the largest pelagic methane reservoirs in the ocean and contribute substantially to open ocean methane cycling. The observed and predicted future expansion of OMZs may therefore considerably affect ocean nutrient and greenhouse gas budgets, and the distributions of oxygen-dependent organisms.[64][65][66][1]
The top 50 cm of deep-sea sediments contains ~1 × 1029 microorganisms,[3][8] and the total abundances of archaea and bacteria in these sediments increase with latitude (from 34° N to 79° N) with specific taxa (such as Marine Group I Thaumarchaeota) contributing disproportionately to the increase.[67] Benthic microorganisms show biogeographic patterns and respond to variations in the quantity and quality of the particulate matter sinking to the seafloor.[68] As a result, climate change is expected to particularly affect the functional processes that deep-sea benthic archaea perform (such as ammonia oxidation) and associated biogeochemical cycles.[67][1]
Aerosols affect cloud formation, thereby influencing sunlight irradiation and precipitation, but the extent to which and the manner in which they influence climate remains uncertain.[69] Marine aerosols consist of a complex mixture of sea salt, non-sea-salt sulfate and organic molecules and can function as nuclei for cloud condensation, influencing the radiation balance and, hence, climate.[70][71] For example, biogenic aerosols in remote marine environments (for example, the Southern Ocean) can increase the number and size of cloud droplets, having similar effects on climate as aerosols in highly polluted regions.[71][72][73][74][75] Specifically, phytoplankton emit dimethylsulfide, and its derivate sulfate promotes cloud condensation.[70][75] Understanding the ways in which marine phytoplankton contribute to aerosols will allow better predictions of how changing ocean conditions will affect clouds and feed back on climate84. In addition, the atmosphere itself contains ~1022 microbial cells, and determining the ability of atmospheric microorganisms to grow and form aggregates will be valuable for assessing their influence on climate.[3][1]
Vegetated coastal habitats are important for carbon sequestration, determined by the full trophic spectrum from predators to herbivores, to plants and their associated microbial communities.[76] Human activity, including anthropogenic climate change, has reduced these habitats over the past 50 years by 25–50%, and the abundance of marine predators has dropped by up to 90%.[76][77][78] Given such extensive perturbation, the effects on microbial communities need to be evaluated because microbial activity determines how much carbon is remineralized and released as CO2 and CH4.[1]
- Climate change affects microorganisms
Climate change perturbs interactions between species and forces species to adapt, migrate and be replaced by others or go extinct28,88. Ocean warming, acidification, eutrophication and overuse (for example, fishing, tourism) together cause the decline of coral reefs and may cause ecosystems shifts towards macroalgae89,90,91,92,93 and benthic cyanobacterial mats94,95. The capacity for corals to adapt to climate change is strongly influenced by the responses of their associated microorganisms, including microalgal symbionts and bacteria96,97,98. The hundreds to thousands of microbial species that live on corals are crucial for host health, for example by recycling the waste products, by provisioning essential nutrients and vitamins and by assisting the immune system to fight pathogens99. However, environmental perturbation or coral bleaching can change the coral microbiome rapidly. Such shifts undoubtedly influence the ecological functions and stability of the coral–microorganism system, potentially affecting the capacity and pace at which corals adapt to climate change, and the relationships between corals and other components of the reef ecosystem99,100.[1]
Generally, microorganisms can disperse more easily than macroscopic organisms. Nevertheless, biogeographic distinctions occur for many microbial species, with dispersal, lifestyle (for example, host association) and environmental factors strongly influencing community composition and function54,101,102,103. Ocean currents and thermal and latitudinal gradients are particularly important for marine communities104,105. If movement to more favourable environments is impossible, evolutionary change may be the only survival mechanism88. Microorganisms, such as bacteria, archaea and microalgae, with large population sizes and rapid asexual generation times have high adaptive potential22. Relatively few studies have examined evolutionary adaptation to ocean acidification or other climate change-relevant environmental variables22,28. Similarly, there is limited understanding of the molecular mechanisms of physiological responses and the implications of those responses for biogeochemical cycles.18,[1]
However, several studies have demonstrated effects of elevated CO2 levels on individual phytoplankton species, which may disrupt broader ecosystem-level processes. A field experiment demonstrated that increasing CO2 levels provide a selective advantage to a toxic microalga, Vicicitus globosus, leading to disruption of organic matter transfer across trophic levels106. The marine cyanobacterial genus Trichodesmium responds to long-term (4.5-year) exposure to elevated CO2 levels with irreversible genetic changes that increase nitrogen fixation and growth107. For the photosynthetic green alga Ostreococcus tauri, elevated CO2 levels increase growth, cell size and carbon-to-nitrogen ratios108. Higher CO2 levels also affect the population structure of O. tauri, with changes in ecotypes and niche occupation, thereby affecting the broader food webs and biogeochemical cycles108. Rather than producing larger cells, the calcifying phytoplankton species Emiliania huxleyi responds to the combined effects of elevated temperature and elevated CO2 levels (and associated acidification) by producing smaller cells that contain less carbon109. However, for this species, overall production rates do not change as a result of evolutionary adaptation to higher CO2 levels109. Responses to CO2 levels differ between communities (for example, between Arctic phytoplankton and Antarctic phytoplankton110). A mesocosm study identified variable changes in the diversity of viruses that infect E. huxleyi when it is growing under elevated CO2 levels, and noted the need to determine whether elevated CO2 levels directly affected viruses, hosts or the interactions between them111. These examples illustrate the need to improve our understanding of evolutionary processes and incorporate that knowledge into predictions of the effects of climate change.[1]
Ocean acidification presents marine microorganisms with pH conditions well outside their recent historical range, which affects their intracellular pH homeostasis18,112. Species that are less adept at regulating internal pH will be more affected, and factors such as organism size, aggregation state, metabolic activity and growth rate influence the capacity for regulation112.[1]
Lower pH causes bacteria and archaea to change gene expression in ways that support cell maintenance rather than growth18. In mesocosms with low phytoplankton biomass, bacteria committed more resources to pH homeostasis than bacteria in nutrient-enriched mesocosms with high phytoplankton biomass. Consequently, ocean acidification is predicted to alter the microbial food web via changes in cellular growth efficiency, carbon cycling and energy fluxes, with the biggest effects expected in the oligotrophic regions, which include most of the ocean18. Experimental comparisons of Synechococcus sp. growth under both present and predicted future pH concentrations showed effects not only on the cyanobacteria but also on the cyanophage viruses that infect them113.[1]
Environmental temperature and latitude correlate with the diversity, distribution and/or temperature optimum (Topt) of certain marine taxa, with models predicting that rising temperatures will cause a poleward shift of cold-adapted communities52,114,115,116,117,118. However, Topt of phytoplankton from polar and temperate waters was found to be substantially higher than environmental temperatures, and an eco-evolutionary model predicted that Topt for tropical phytoplankton would be substantially higher than observed experimental values116. Understanding how well microorganisms are adapted to environmental temperature and predicting how they will respond to warming requires assessments of more than Topt, which is generally a poor indicator of physiological and ecological adaptation of microorganisms from cold environments119.[1]
Many environmental and physiological factors influence the responses and overall competitiveness of microorganisms in their native environment. For example, elevated temperatures increase protein synthesis in eukaryotic phytoplankton while reducing cellular ribosome concentration120. As the biomass of eukaryotic phytoplankton is ~1 Gt C (ref.13) and ribosomes are phosphate rich, climate change-driven alteration of their nitrogen-to-phosphate ratio will affect resource allocation in the global ocean120. Ocean warming is thought to favour smaller plankton types over larger ones, changing biogeochemical fluxes such as particle export121. Increased ocean temperatures, acidification and decreased nutrient supplies are projected to increase the extracellular release of dissolved organic matter from phytoplankton, with changes in the microbial loop possibly causing increased microbial production at the expense of higher trophic levels122. Warming can also alleviate iron limitation of nitrogen-fixing cyanobacteria, with potentially profound implications for new nitrogen supplied to food webs of the future warming oceans123. Careful attention needs to be paid to how to quantify and interpret responses of environmental microorganisms to ecosystem changes and stresses linked to climate change124,125. Key questions thus remain about the functional consequences of community shifts, such as changes in carbon remineralization versus carbon sequestration, and nutrient cycling.[1]
Terrestrial biome
[edit]There is ~100-fold more terrestrial biomass than marine biomass, and terrestrial plants account for a large proportion of Earth’s total biomass15. Terrestrial plants perform roughly half of net global primary production30,67. Soils store ~2,000 billion tonnes of organic carbon, which is more than the combined pool of carbon in the atmosphere and vegetation126. The total number of microorganisms in terrestrial environments is ~1029, similar to the total number in marine environments8. Soil microorganisms regulate the amount of organic carbon stored in soil and released back to the atmosphere, and indirectly influence carbon storage in plants and soils through provision of macronutrients that regulate productivity (nitrogen and phosphorus)126,127. Plants provide a substantial amount of carbon to their mycorrhizal fungal symbionts, and in many ecosystems, mycorrhizal fungi are responsible for substantial amounts of nitrogen and phosphorus acquisition by plants128.[1]
Plants remove CO2 from the atmosphere through photosynthesis and create organic matter that fuels terrestrial ecosystems. Conversely, autotrophic respiration by plants (60 Pg C per year) and heterotrophic respiration by microorganisms (60 Pg C per year) release CO2 back into the atmosphere126,129. Temperature influences the balance between these opposing processes and thus the capacity of the terrestrial biosphere to capture and store anthropogenic carbon emissions (currently, storing approximately one quarter of emissions) (Fig. 1). Warming is expected to accelerate carbon release into the atmosphere129.[1]
Forests cover ~30% of the land surface, contain ~45% of terrestrial carbon, make up ~50% of terrestrial primary production and sequester up to 25% of anthropogenic CO2 (refs130,131). Grasslands cover ~29% of the terrestrial surface132. Non-forested, arid and semiarid regions (47%) are important for the carbon budget and respond differently to anthropogenic climate change than forested regions132,133. Lakes make up ~4% of the non-glaciated land area134, and shallow lakes emit substantial amounts of CH4 (refs135,136). Peat (decomposed plant litter) covers ~3% of the land surface and, due to plant productivity exceeding decomposition, intact peatlands function as a global carbon sink and contain ~30% of global soil carbon137,138. In permafrost, the accumulation of carbon in organic matter (remnants of plants, animals and microorganisms) far exceeds the respiratory losses, creating the largest terrestrial carbon sink139,140,141. Climate warming of 1.5–2 °C (relative to the global mean surface temperature in 1850–1900) is predicted to reduce permafrost by 28–53% (compared with levels in 1960–1990)142, thereby making large carbon reservoirs available for microbial respiration and greenhouse gas emissions.[1]
Evaluations of the top 10 cm of soil143 and whole-soil profiles to 100 cm deep, which contain older stocks of carbon144, demonstrate that warming increases carbon loss to the atmosphere. Explaining differences in carbon loss between different soil sites will require a greater range of predictive variables (in addition to soil organic matter content, temperature, precipitation, pH and clay content)145,146. Nevertheless, predictions from global assessments of responses to warming indicate that terrestrial carbon loss under warming is causing a positive feedback that will accelerate the rate of climate change143, particularly in cold and temperate soils, which store much of the global soil carbon147.[1]
- Microorganisms affect climate change
Higher CO2 levels in the atmosphere increase primary productivity and thus forest leaf and root litter148,149,150, which leads to higher carbon emissions due to microbial degradation151. Higher temperatures promote higher rates of terrestrial organic matter decomposition152. The effect of temperature is not just a kinetic effect on microbial reaction rates but results from plant inputs stimulating microbial growth152,153,154.[1]
Several local environmental factors (such as microbial community composition, density of dead wood, nitrogen availability and moisture) influence rates of microbial activity (for example, fungal colonization of wood) necessitating Earth system model predictions of soil carbon losses through climate warming to incorporate local controls on ecosystem processes155. In this regard, plant nutrient availability affects the net carbon balance in forests, with nutrient-poor forests releasing more carbon than nutrient-rich forests156. Microbial respiration may be lower in nutrient-rich forests as plants provide less carbon (for example, as root exudates) to rhizosphere microorganisms157.[1]
Plants release ~50% of fixed carbon into the soil, which is available for microbial growth158,159,160. In addition to microorganisms using exudates as an energy source, exudates can disrupt mineral–organic associations, liberating organic compounds from minerals that are used for microbial respiration, thereby increasing carbon release159. The relevance of these plant–mineral interactions illustrates the importance of biotic–abiotic interactions, in addition to biotic interactions (plant–microorganism) when one is evaluating the influence of climate change159. Thermodynamic models that incorporate the interactions of microorganisms and secreted enzymes with organic matter and minerals have been used to predict soil carbon–climate feedbacks in response to increasing temperature; one study predicted more variable but weaker soil carbon–climate feedbacks from a thermodynamic model than from static models160.[1]
The availability of soil organic matter for microbial degradation versus long-term storage depends on many environmental factors, including the soil mineral characteristics, acidity and redox state; water availability; climate; and the types of microorganisms present in the soil161. The nature of the organic matter, in particular substrate complexity, affects microbial decomposition. Furthermore, the microbial capacity to access organic matter differs between soil types (for example, with different clay content)162. If access is taken into account, increasing atmospheric CO2 levels are predicted to allow greater microbial decomposition and less soil retention of organic carbon162.[1]
Elevated CO2 concentrations enhance competition for nitrogen between plants and microorganisms163. Herbivores (invertebrates and mammals) affect the amount of organic matter that is returned to soil and thereby microbial biomass and activity164. For example, grasshoppers diminish plant biomass and plant nitrogen demand, thereby increasing microbial activity163. Climate change can reduce herbivory, resulting in overall alterations to global nitrogen and carbon cycles that reduce terrestrial carbon sequestration163. Detritivores (for example, earthworms) influence greenhouse gas emissions by indirectly affecting plants (for example, by increasing soil fertility) and soil microorganisms165. Earthworms modify soils through feeding, burrowing and deposition of waste products. The anaerobic gut environment of earthworms harbours microorganisms that perform denitrification and produce N2O. Earthworms enhance soil fertility, and their presence can result in net greenhouse gas emissions165, although the combined effects of increased temperature and decreased rainfall on detritivore feeding and microbial respiration may reduce emissions166.[1]
In peatlands, decay-resistant litter (for example, antimicrobial phenolics and polysaccharides of Sphagnum mosses) inhibits microbial decomposition, and water saturation restricts oxygen exchange and promotes the growth of anaerobes and release of CO2 and CH4 (refs137,167). Increased temperature and reduced soil water content caused by climate change promote the growth of vascular plants (ericaceous shrubs) but reduce the productivity of peat moss. Changes in plant litter composition and associated microbial processes (for example, reduced immobilization of nitrogen and enhanced heterotrophic respiration) are switching peatlands from carbon sinks to carbon sources137.[1]
Melting and degradation of permafrost allows microbial decomposition of previously frozen carbon, releasing CO2 and CH4 (refs139,140,141,168,169). Coastal permafrost erosion will lead to the mobilization of large quantities of carbon to the ocean, with potentially large CO2 emissions occurring through increased microbial remineralization170, causing a positive feedback loop that accelerates climate change139,140,141,168,169,170,171. Melting of permafrost leads to increases in water-saturated soils172, which promotes anaerobic CH4 production by methanogens and CO2 production by a range of microorganisms. Production is slow compared with metabolism in drained aerobic soils, which release CO2 rather than CH4. However, a 7-year laboratory study of CO2 and CH4 production found that once methanogen communities became active in thawing permafrost, equal amounts of CO2 and CH4 were formed under anoxic conditions, and it was predicted that by the end of the century, carbon emissions from anoxic environments will drive climate change to a greater extent than emissions from oxic environments172.[1]
A 15-year mesocosm study that simulated freshwater lake environments determined that the combined effects of eutrophication and warming can lead to large increases in CH4 ebullition (bubbles from accumulated gas)135. As small lakes are susceptible to eutrophication and tend to be located in climate-sensitive regions, the role of lake microorganisms in contributing to global greenhouse gas emissions needs to be evaluated135,136.[1]
- Climate change affects microorganisms
Shifts in climate can influence the structure and diversity of microbial communities directly (for example, seasonality and temperature) or indirectly (for example, plant composition, plant litter and root exudates). Soil microbial diversity influences plant diversity and is important for ecosystem functions, including carbon cycling173,174.[1]
Both short-term laboratory warming and long-term (more than 50 years) natural geothermal warming initially increased the growth and respiration of soil microorganisms, leading to net CO2 release and subsequent depletion of substrates, causing a decrease in biomass and reduced microbial activity175. This implies that microbial communities do not readily adapt to higher temperatures, and the resulting effects on reaction rates and substrate depletion reduce overall carbon loss175. By contrast, a 10-year study found that soil communities adapted to increased temperature by changing composition and patterns of substrate use, leading to less carbon loss than would have occurred without adaptation176. Substantial changes in bacterial and fungal communities were also found in forest soils with a more than 20 °C average annual temperature range177, and in response to warming across a 9-year study of tall-grass prairie soils178.[1]
Two studies assessed the effects of elevated temperatures on microbial respiration rates and mechanisms and outcomes of adaptation179,180. The studies examined a wide range of environmental temperatures (−2 to 28 °C), dryland soils (110 samples) and boreal, temperate and tropical soils (22 samples), and evaluated how communities respond to three different temperatures (~10–30 °C). Thermal adaptation was linked to biophysical characteristics of cell membranes and enzymes (reflecting activity-stability trade-offs180) and the genomic potential of microorganisms (with warmer environments having microbial communities with more diverse lifestyles179). Respiration rates per unit biomass were lower in soils from higher-temperature environments, indicating that thermal adaptation of microbial communities may lessen positive climate feedbacks. However, as respiration depends on multiple interrelated factors (not just on one variable, such as temperature), such mechanistic insights into microbial physiology need to be represented in biogeochemical models of possible positive climate feedbacks.[1]
Microbial growth responses to temperature change are complex and varied181. Microbial growth efficiency is a measure of how effectively microorganisms convert organic matter into biomass, with lower efficiency meaning more carbon is released to the atmosphere182,183. A 1-week laboratory study found that increasing temperature led to increases in microbial turnover but no change in microbial growth efficiency, and predicted that warming would promote carbon accumulation in soil183. A field study spanning 18 years found microbial efficiency was reduced at higher soil temperature, with decomposition of recalcitrant, complex substrates increasing by the end of the period along with a net loss of soil carbon182.[1]
Similarly, in a 26-year forest-soil warming study, temporal variation occurred in organic matter decomposition and CO2 release184, leading to changes in microbial community composition and carbon use efficiency, reduced microbial biomass and reduced microbially accessible carbon184. Overall, the study predicted anthropogenic climate change to cause long-term, increasing and sustained carbon release184. Similar predictions arise from Earth system models that simulate microbial physiological responses185 or incorporate the effects of freezing and thawing of cold-climate soils186.[1]
Climate change directly and indirectly influences microbial communities and their functions through several interrelated factors, such as temperature, precipitation, soil properties and plant input. As soil microorganisms in deserts are carbon limited, increased carbon input from plants promotes transformation of nitrogenous compounds, microbial biomass, diversity (for example, of fungi), enzymatic activity and use of recalcitrant organic matter133. Although these changes may enhance respiration and net loss of carbon from soil, the specific characteristics of arid and semiarid regions may mean they could function as carbon sinks133. However, a study of 19 temperate grassland sites found that seasonal differences in rainfall constrain biomass accumulation132. To better understand aboveground plant-biomass responses to CO2 levels and seasonal precipitation, we also need improved knowledge of microbial community responses and functions.[1]
Metagenome data, including metagenome-assembled genomes, provide knowledge of key microbial groups that metabolize organic matter and release CO2 and CH4 and link these groups to the biogeochemistry occurring in thawing permafrost187,188,189,190,191. Tundra microbial communities change in the soil layer of permafrost after warming192. Within 1.5 years of warming, the functional potential of the microbial communities changed markedly, with an increasing abundance of genes involved in aerobic and anaerobic carbon decomposition and nutrient cycling. Although microbial metabolism stimulates primary productivity by plants, the balance between microbial respiration and primary productivity results in a net release of carbon to the atmosphere192. When forests expand into warming regions of tundra, plant growth can produce a net loss of carbon, possibly as a result of root exudates stimulating microbial decomposition of native soil carbon153,193. Although there are reports of carbon accumulating owing to warming (for example, ref.183), most studies describe microbial community responses that result in carbon loss.[1]
Rapid warming of the Antarctic Peninsula and associated islands resulted in range expansion of Antarctic hair grass (Deschampsia antarctica), as it outcompetes other indigenous species (for example, the moss Sanionia uncinata) through the superior capacity of its roots to acquire peptides and thus nitrogen194. The ability of the grass to be competitive depends on microbial digestion of extracellular proteins and generation of amino acids, nitrate and ammonium194. As warmer soils in this region harbour greater fungal diversity, climate change is predicted to cause changes in the fungal communities that will affect nutrient cycling and primary productivity195. Cyanobacterial diversity and toxin production within benthic mats from both the Antarctic Peninsula and the Arctic increased during 6 months of exposure to high growth temperatures196. A shift to toxin-producing species or increased toxin production by existing species could affect polar freshwater lakes, where cyanobacteria are often the dominant benthic primary producers196.[1]
Conclusion
[edit]Microorganisms make a major contribution to carbon sequestration, particularly marine phytoplankton, which fix as much net CO2 as terrestrial plants. For this reason, environmental changes that affect marine microbial photosynthesis and subsequent storage of fixed carbon in deep waters are of major importance for the global carbon cycle. Microorganisms also contribute substantially to greenhouse gas emissions via heterotrophic respiration (CO2), methanogenesis (CH4) and denitrification (N2O).[1]
Many factors influence the balance of microbial greenhouse gas capture versus emission, including the biome, the local environment, food web interactions and responses, and particularly anthropogenic climate change and other human activities (Figs 1–3).[1][1]
Human activity that directly affects microorganisms includes greenhouse gas emissions (particularly CO2, CH4 and N2O), pollution (particularly eutrophication), agriculture (particularly land usage) and population growth, which positively feeds back on climate change, pollution, agricultural practice and the spread of disease. Human activity that alters the ratio of carbon uptake relative to release will drive positive feedbacks and accelerate the rate of climate change. By contrast, microorganisms also offer important opportunities for remedying human-caused problems through improved agricultural outcomes, production of biofuels and remediation of pollution.[1]
References
[edit]- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as Cavicchioli, Ricardo; Ripple, William J.; Timmis, Kenneth N.; Azam, Farooq; Bakken, Lars R.; Baylis, Matthew; Behrenfeld, Michael J.; Boetius, Antje; Boyd, Philip W.; Classen, Aimée T.; Crowther, Thomas W.; Danovaro, Roberto; Foreman, Christine M.; Huisman, Jef; Hutchins, David A.; Jansson, Janet K.; Karl, David M.; Koskella, Britt; Mark Welch, David B.; Martiny, Jennifer B. H.; Moran, Mary Ann; Orphan, Victoria J.; Reay, David S.; Remais, Justin V.; Rich, Virginia I.; Singh, Brajesh K.; Stein, Lisa Y.; Stewart, Frank J.; Sullivan, Matthew B.; et al. (2019). "Scientists' warning to humanity: Microorganisms and climate change". Nature Reviews Microbiology. 17 (9): 569–586. doi:10.1038/s41579-019-0222-5. PMC 7136171. PMID 31213707.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Cite error: The named reference "Cavicchioli2019" was defined multiple times with different content (see the help page).
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. Cite error: The named reference "Cavicchioli2019" was defined multiple times with different content (see the help page).
- ^ United Nations (2018) The Sustainable Development Goals Report 2018, United Nations Department of Economic and Social Affairs.
- ^ a b c Flemming, Hans-Curt; Wuertz, Stefan (2019). "Bacteria and archaea on Earth and their abundance in biofilms". Nature Reviews Microbiology. 17 (4): 247–260. doi:10.1038/s41579-019-0158-9. PMID 30760902. S2CID 61155774.
- ^ Karner, Markus B.; Delong, Edward F.; Karl, David M. (2001). "Archaeal dominance in the mesopelagic zone of the Pacific Ocean". Nature. 409 (6819): 507–510. Bibcode:2001Natur.409..507K. doi:10.1038/35054051. PMID 11206545. S2CID 6789859.
- ^ a b Azam, Farooq; Malfatti, Francesca (2007). "Microbial structuring of marine ecosystems". Nature Reviews Microbiology. 5 (10): 782–791. doi:10.1038/nrmicro1747. PMID 17853906. S2CID 10055219.
- ^ Kallmeyer, J.; Pockalny, R.; Adhikari, R. R.; Smith, D. C.; d'Hondt, S. (2012). "Global distribution of microbial abundance and biomass in subseafloor sediment". Proceedings of the National Academy of Sciences. 109 (40): 16213–16216. doi:10.1073/pnas.1203849109. PMC 3479597. PMID 22927371.
- ^ Bar-On, Yinon M.; Phillips, Rob; Milo, Ron (2018). "The biomass distribution on Earth". Proceedings of the National Academy of Sciences. 115 (25): 6506–6511. doi:10.1073/pnas.1711842115. PMC 6016768. PMID 29784790.
- ^ a b Danovaro, R.; Corinaldesi, C.; Rastelli, E.; Dell'Anno, A. (2015). "Towards a better quantitative assessment of the relevance of deep-sea viruses, Bacteria and Archaea in the functioning of the ocean seafloor". Aquatic Microbial Ecology. 75: 81–90. doi:10.3354/ame01747.
- ^ Caldeira, Ken; Wickett, Michael E. (2003). "Anthropogenic carbon and ocean pH". Nature. 425 (6956): 365. doi:10.1038/425365a. PMID 14508477. S2CID 4417880.
- ^ a b Bunse, Carina; Lundin, Daniel; Karlsson, Christofer M. G.; Akram, Neelam; Vila-Costa, Maria; Palovaara, Joakim; Svensson, Lovisa; Holmfeldt, Karin; González, José M.; Calvo, Eva; Pelejero, Carles; Marrasé, Cèlia; Dopson, Mark; Gasol, Josep M.; Pinhassi, Jarone (2016). "Response of marine bacterioplankton pH homeostasis gene expression to elevated CO2". Nature Climate Change. 6 (5): 483–487. Bibcode:2016NatCC...6..483B. doi:10.1038/nclimate2914.
- ^ a b c Hurd, Catriona L.; Lenton, Andrew; Tilbrook, Bronte; Boyd, Philip W. (2018). "Current understanding and challenges for oceans in a higher-CO2 world". Nature Climate Change. 8 (8): 686–694. Bibcode:2018NatCC...8..686H. doi:10.1038/s41558-018-0211-0. S2CID 92793532.
- ^ Honisch, B.; Ridgwell, A.; Schmidt, D. N.; Thomas, E.; Gibbs, S. J.; Sluijs, A.; Zeebe, R.; Kump, L.; Martindale, R. C.; Greene, S. E.; Kiessling, W.; Ries, J.; Zachos, J. C.; Royer, D. L.; Barker, S.; Marchitto, T. M.; Moyer, R.; Pelejero, C.; Ziveri, P.; Foster, G. L.; Williams, B. (2012). "The Geological Record of Ocean Acidification". Science. 335 (6072): 1058–1063. Bibcode:2012Sci...335.1058H. doi:10.1126/science.1208277. hdl:1874/385704. PMID 22383840. S2CID 6361097.
- ^ Sosdian, S.M.; Greenop, R.; Hain, M.P.; Foster, G.L.; Pearson, P.N.; Lear, C.H. (2018). "Constraining the evolution of Neogene ocean carbonate chemistry using the boron isotope pH proxy". Earth and Planetary Science Letters. 498: 362–376. Bibcode:2018E&PSL.498..362S. doi:10.1016/j.epsl.2018.06.017. S2CID 52230107.
- ^ Riebesell, Ulf; Gattuso, Jean-Pierre (2015). "Lessons learned from ocean acidification research". Nature Climate Change. 5 (1): 12–14. Bibcode:2015NatCC...5...12R. doi:10.1038/nclimate2456.
- ^ Pörtner, H.-O. et al. in: Climate Change 2014 — Impacts, Adaptation and Vulnerability: Part A: Global and Sectoral Aspects: Working Group II Contribution to the IPCC Fifth Assessment Report (eds Field, C. B. et al.) 411–484 (Cambridge University Press, 2014)
- ^ Brennan, Georgina; Collins, Sinéad (2015). "Growth responses of a green alga to multiple environmental drivers". Nature Climate Change. 5 (9): 892–897. Bibcode:2015NatCC...5..892B. doi:10.1038/nclimate2682.
- ^ a b c Hutchins, D. A.; Boyd, P. W. (2016). "Marine phytoplankton and the changing ocean iron cycle". Nature Climate Change. 6 (12): 1072–1079. Bibcode:2016NatCC...6.1072H. doi:10.1038/nclimate3147.
- ^ Cite error: The named reference
Microorganisms and ocean global chawas invoked but never defined (see the help page). - ^ Rintoul, S. R.; Chown, S. L.; Deconto, R. M.; England, M. H.; Fricker, H. A.; Masson-Delmotte, V.; Naish, T. R.; Siegert, M. J.; Xavier, J. C. (2018). "Choosing the future of Antarctica". Nature. 558 (7709): 233–241. Bibcode:2018Natur.558..233R. doi:10.1038/s41586-018-0173-4. hdl:10044/1/60056. PMID 29899481. S2CID 49193026.
- ^ a b c d Behrenfeld, Michael J. (2014). "Climate-mediated dance of the plankton". Nature Climate Change. 4 (10): 880–887. Bibcode:2014NatCC...4..880B. doi:10.1038/nclimate2349.
- ^ De Baar, Hein J. W.; De Jong, Jeroen T. M.; Bakker, Dorothée C. E.; Löscher, Bettina M.; Veth, Cornelis; Bathmann, Uli; Smetacek, Victor (1995). "Importance of iron for plankton blooms and carbon dioxide drawdown in the Southern Ocean". Nature. 373 (6513): 412–415. Bibcode:1995Natur.373..412D. doi:10.1038/373412a0. S2CID 4257465.
- ^ Boyd, P. W.; Jickells, T.; Law, C. S.; Blain, S.; Boyle, E. A.; Buesseler, K. O.; Coale, K. H.; Cullen, J. J.; De Baar, H. J. W.; Follows, M.; Harvey, M.; Lancelot, C.; Levasseur, M.; Owens, N. P. J.; Pollard, R.; Rivkin, R. B.; Sarmiento, J.; Schoemann, V.; Smetacek, V.; Takeda, S.; Tsuda, A.; Turner, S.; Watson, A. J. (2007). "Mesoscale Iron Enrichment Experiments 1993-2005: Synthesis and Future Directions". Science. 315 (5812): 612–617. Bibcode:2007Sci...315..612B. doi:10.1126/science.1131669. PMID 17272712. S2CID 2476669.
- ^ Behrenfeld, Michael J.; o'Malley, Robert T.; Boss, Emmanuel S.; Westberry, Toby K.; Graff, Jason R.; Halsey, Kimberly H.; Milligan, Allen J.; Siegel, David A.; Brown, Matthew B. (2016). "Revaluating ocean warming impacts on global phytoplankton". Nature Climate Change. 6 (3): 323–330. Bibcode:2016NatCC...6..323B. doi:10.1038/nclimate2838.
- ^ a b c Behrenfeld, Michael J.; Hu, Yongxiang; o'Malley, Robert T.; Boss, Emmanuel S.; Hostetler, Chris A.; Siegel, David A.; Sarmiento, Jorge L.; Schulien, Jennifer; Hair, Johnathan W.; Lu, Xiaomei; Rodier, Sharon; Scarino, Amy Jo (2017). "Annual boom–bust cycles of polar phytoplankton biomass revealed by space-based lidar". Nature Geoscience. 10 (2): 118–122. Bibcode:2017NatGe..10..118B. doi:10.1038/ngeo2861.
- ^ Behrenfeld, Michael J.; o'Malley, Robert T.; Siegel, David A.; McClain, Charles R.; Sarmiento, Jorge L.; Feldman, Gene C.; Milligan, Allen J.; Falkowski, Paul G.; Letelier, Ricardo M.; Boss, Emmanuel S. (2006). "Climate-driven trends in contemporary ocean productivity". Nature. 444 (7120): 752–755. Bibcode:2006Natur.444..752B. doi:10.1038/nature05317. PMID 17151666. S2CID 4414391.
- ^ Levitan, O.; Rosenberg, G.; Setlik, I.; Setlikova, E.; Grigel, J.; Klepetar, J.; Prasil, O.; Berman-Frank, I. (2007). "Elevated CO2enhances nitrogen fixation and growth in the marine cyanobacterium Trichodesmium". Global Change Biology. 13 (2): 531–538. Bibcode:2007GCBio..13..531L. doi:10.1111/j.1365-2486.2006.01314.x. S2CID 86121269.
- ^ Verspagen, Jolanda M. H.; Van De Waal, Dedmer B.; Finke, Jan F.; Visser, Petra M.; Huisman, Jef (2014). "Contrasting effects of rising CO2on primary production and ecological stoichiometry at different nutrient levels". Ecology Letters. 17 (8): 951–960. doi:10.1111/ele.12298. hdl:20.500.11755/ecac2c45-7efa-4c90-9e29-f2bafcee1c95. PMID 24813339.
- ^ Holding, J. M.; Duarte, C. M.; Sanz-Martín, M.; Mesa, E.; Arrieta, J. M.; Chierici, M.; Hendriks, I. E.; García-Corral, L. S.; Regaudie-De-Gioux, A.; Delgado, A.; Reigstad, M.; Wassmann, P.; Agustí, S. (2015). "Temperature dependence of CO2-enhanced primary production in the European Arctic Ocean". Nature Climate Change. 5 (12): 1079–1082. Bibcode:2015NatCC...5.1079H. doi:10.1038/nclimate2768. hdl:10754/596052.
- ^ Boyce, Daniel G.; Lewis, Marlon R.; Worm, Boris (2010). "Global phytoplankton decline over the past century". Nature. 466 (7306): 591–596. Bibcode:2010Natur.466..591B. doi:10.1038/nature09268. PMID 20671703. S2CID 2413382.
- ^ MacKas, David L. (2011). "Does blending of chlorophyll data bias temporal trend?". Nature. 472 (7342): E4–E5. Bibcode:2011Natur.472E...4M. doi:10.1038/nature09951. PMID 21490623. S2CID 4308744.
- ^ Rykaczewski, Ryan R.; Dunne, John P. (2011). "A measured look at ocean chlorophyll trends". Nature. 472 (7342): E5–E6. Bibcode:2011Natur.472E...5R. doi:10.1038/nature09952. PMID 21490624. S2CID 205224535.
- ^ McQuatters-Gollop, Abigail; Reid, Philip C.; Edwards, Martin; Burkill, Peter H.; Castellani, Claudia; Batten, Sonia; Gieskes, Winfried; Beare, Doug; Bidigare, Robert R.; Head, Erica; Johnson, Rod; Kahru, Mati; Koslow, J. Anthony; Pena, Angelica (2011). "Is there a decline in marine phytoplankton?". Nature. 472 (7342): E6–E7. Bibcode:2011Natur.472E...6M. doi:10.1038/nature09950. PMID 21490625. S2CID 205224519.
- ^ Boyce, Daniel G.; Lewis, Marlon R.; Worm, Boris (2011). "Boyce et al. Reply". Nature. 472 (7342): E8–E9. Bibcode:2011Natur.472E...8B. doi:10.1038/nature09953. S2CID 4317554.
- ^ Antoine, David; Morel, André; Gordon, Howard R.; Banzon, Viva F.; Evans, Robert H. (2005). "Bridging ocean color observations of the 1980s and 2000s in search of long-term trends". Journal of Geophysical Research. 110 (C6): C06009. Bibcode:2005JGRC..110.6009A. doi:10.1029/2004JC002620.
- ^ Wernand, Marcel R.; Van Der Woerd, Hendrik J.; Gieskes, Winfried W. C. (2013). "Trends in Ocean Colour and Chlorophyll Concentration from 1889 to 2000, Worldwide". PLOS ONE. 8 (6): e63766. Bibcode:2013PLoSO...863766W. doi:10.1371/journal.pone.0063766. PMC 3680421. PMID 23776435.
- ^ a b Rousseaux, Cecile S.; Gregg, Watson W. (2015). "Recent decadal trends in global phytoplankton composition". Global Biogeochemical Cycles. 29 (10): 1674–1688. Bibcode:2015GBioC..29.1674R. doi:10.1002/2015GB005139. S2CID 130686652.
- ^ Kirchman, David L.; Morán, Xosé Anxelu G.; Ducklow, Hugh (2009). "Microbial growth in the polar oceans — role of temperature and potential impact of climate change". Nature Reviews Microbiology. 7 (6): 451–459. doi:10.1038/nrmicro2115. PMID 19421189. S2CID 31230080.
- ^ Dore, J. E.; Lukas, R.; Sadler, D. W.; Church, M. J.; Karl, D. M. (2009). "Physical and biogeochemical modulation of ocean acidification in the central North Pacific". Proceedings of the National Academy of Sciences. 106 (30): 12235–12240. doi:10.1073/pnas.0906044106. PMC 2716384. PMID 19666624.
- ^ Saba, Vincent S.; Friedrichs, Marjorie A. M.; Carr, Mary-Elena; Antoine, David; Armstrong, Robert A.; Asanuma, Ichio; Aumont, Olivier; Bates, Nicholas R.; Behrenfeld, Michael J.; Bennington, Val; Bopp, Laurent; Bruggeman, Jorn; Buitenhuis, Erik T.; Church, Matthew J.; Ciotti, Aurea M.; Doney, Scott C.; Dowell, Mark; Dunne, John; Dutkiewicz, Stephanie; Gregg, Watson; Hoepffner, Nicolas; Hyde, Kimberly J. W.; Ishizaka, Joji; Kameda, Takahiko; Karl, David M.; Lima, Ivan; Lomas, Michael W.; Marra, John; McKinley, Galen A.; et al. (2010). "Challenges of modeling depth-integrated marine primary productivity over multiple decades: A case study at BATS and HOT". Global Biogeochemical Cycles. 24 (3): n/a. Bibcode:2010GBioC..24.3020S. doi:10.1029/2009GB003655. S2CID 21669863.
- ^ a b Buttigieg, Pier Luigi; Fadeev, Eduard; Bienhold, Christina; Hehemann, Laura; Offre, Pierre; Boetius, Antje (2018). "Marine microbes in 4D — using time series observation to assess the dynamics of the ocean microbiome and its links to ocean health". Current Opinion in Microbiology. 43: 169–185. doi:10.1016/j.mib.2018.01.015. PMID 29477022. S2CID 3528036.
- ^ Rusch, Douglas B.; Halpern, Aaron L.; Sutton, Granger; Heidelberg, Karla B.; Williamson, Shannon; Yooseph, Shibu; Wu, Dongying; Eisen, Jonathan A.; Hoffman, Jeff M.; Remington, Karin; Beeson, Karen; Tran, Bao; Smith, Hamilton; Baden-Tillson, Holly; Stewart, Clare; Thorpe, Joyce; Freeman, Jason; Andrews-Pfannkoch, Cynthia; Venter, Joseph E.; Li, Kelvin; Kravitz, Saul; Heidelberg, John F.; Utterback, Terry; Rogers, Yu-Hui; Falcón, Luisa I.; Souza, Valeria; Bonilla-Rosso, Germán; Eguiarte, Luis E.; Karl, David M.; et al. (2007). "The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific". PLOS Biology. 5 (3): e77. doi:10.1371/journal.pbio.0050077. PMC 1821060. PMID 17355176.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Brown, Mark V.; Lauro, Federico M.; Demaere, Matthew Z.; Muir, Les; Wilkins, David; Thomas, Torsten; Riddle, Martin J.; Fuhrman, Jed A.; Andrews‐Pfannkoch, Cynthia; Hoffman, Jeffrey M.; McQuaid, Jeffrey B.; Allen, Andrew; Rintoul, Stephen R.; Cavicchioli, Ricardo (2012). "Global biogeography of SAR11 marine bacteria". Molecular Systems Biology. 8: 595. doi:10.1038/msb.2012.28. PMC 3421443. PMID 22806143. S2CID 15711981.
- ^ Wilkins, David; Lauro, Federico M.; Williams, Timothy J.; Demaere, Matthew Z.; Brown, Mark V.; Hoffman, Jeffrey M.; Andrews-Pfannkoch, Cynthia; McQuaid, Jeffrey B.; Riddle, Martin J.; Rintoul, Stephen R.; Cavicchioli, Ricardo (2013). "Biogeographic partitioning of Southern Ocean microorganisms revealed by metagenomics". Environmental Microbiology. 15 (5): 1318–1333. doi:10.1111/1462-2920.12035. PMID 23199136.
- ^ Sunagawa, S.; Coelho, L. P.; Chaffron, S.; Kultima, J. R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D. R.; Alberti, A.; Cornejo-Castillo, F. M.; Costea, P. I.; Cruaud, C.; d'Ovidio, F.; Engelen, S.; Ferrera, I.; Gasol, J. M.; Guidi, L.; Hildebrand, F.; Kokoszka, F.; Lepoivre, C.; Lima-Mendez, G.; Poulain, J.; Poulos, B. T.; Royo-Llonch, M.; Sarmento, H.; Vieira-Silva, S.; Dimier, C.; Picheral, M.; et al. (2015). "Structure and function of the global ocean microbiome". Science. 348 (6237). doi:10.1126/science.1261359. hdl:10261/117712. PMID 25999513. S2CID 206562917.
- ^ Brum, J. R.; Ignacio-Espinoza, J. C.; Roux, S.; Doulcier, G.; Acinas, S. G.; Alberti, A.; Chaffron, S.; Cruaud, C.; De Vargas, C.; Gasol, J. M.; Gorsky, G.; Gregory, A. C.; Guidi, L.; Hingamp, P.; Iudicone, D.; Not, F.; Ogata, H.; Pesant, S.; Poulos, B. T.; Schwenck, S. M.; Speich, S.; Dimier, C.; Kandels-Lewis, S.; Picheral, M.; Searson, S.; Bork, P.; Bowler, C.; Sunagawa, S.; Wincker, P.; et al. (2015). "Patterns and ecological drivers of ocean viral communities". Science. 348 (6237). doi:10.1126/science.1261498. hdl:10261/117735. PMID 25999515. S2CID 206563008.
- ^ De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahe, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; Carmichael, M.; Poulain, J.; Romac, S.; Colin, S.; Aury, J.-M.; Bittner, L.; Chaffron, S.; Dunthorn, M.; Engelen, S.; Flegontova, O.; Guidi, L.; Horak, A.; Jaillon, O.; Lima-Mendez, G.; Luke, J.; Malviya, S.; Morard, R.; Mulot, M.; Scalco, E.; et al. (2015). "Eukaryotic plankton diversity in the sunlit ocean" (PDF). Science. 348 (6237). doi:10.1126/science.1261605. PMID 25999516. S2CID 12853481.
- ^ Lima-Mendez, G.; Faust, K.; Henry, N.; Decelle, J.; Colin, S.; Carcillo, F.; Chaffron, S.; Ignacio-Espinosa, J. C.; Roux, S.; Vincent, F.; Bittner, L.; Darzi, Y.; Wang, J.; Audic, S.; Berline, L.; Bontempi, G.; Cabello, A. M.; Coppola, L.; Cornejo-Castillo, F. M.; d'Ovidio, F.; De Meester, L.; Ferrera, I.; Garet-Delmas, M.-J.; Guidi, L.; Lara, E.; Pesant, S.; Royo-Llonch, M.; Salazar, G.; Sanchez, P.; et al. (2015). "Determinants of community structure in the global plankton interactome". Science. 348 (6237). doi:10.1126/science.1262073. hdl:10261/117702. PMID 25999517. S2CID 10326640.
- ^ a b Guidi, Lionel; Chaffron, Samuel; Bittner, Lucie; Eveillard, Damien; Larhlimi, Abdelhalim; Roux, Simon; Darzi, Youssef; Audic, Stephane; Berline, Léo; Brum, Jennifer R.; Coelho, Luis Pedro; Espinoza, Julio Cesar Ignacio; Malviya, Shruti; Sunagawa, Shinichi; Dimier, Céline; Kandels-Lewis, Stefanie; Picheral, Marc; Poulain, Julie; Searson, Sarah; Stemmann, Lars; Not, Fabrice; Hingamp, Pascal; Speich, Sabrina; Follows, Mick; Karp-Boss, Lee; Boss, Emmanuel; Ogata, Hiroyuki; Pesant, Stephane; Weissenbach, Jean; et al. (2016). "Plankton networks driving carbon export in the oligotrophic ocean". Nature. 532 (7600): 465–470. Bibcode:2016Natur.532..465.. doi:10.1038/nature16942. PMC 4851848. PMID 26863193.
- ^ Roux, Simon; Brum, Jennifer R.; Dutilh, Bas E.; Sunagawa, Shinichi; Duhaime, Melissa B.; Loy, Alexander; Poulos, Bonnie T.; Solonenko, Natalie; Lara, Elena; Poulain, Julie; Pesant, Stéphane; Kandels-Lewis, Stefanie; Dimier, Céline; Picheral, Marc; Searson, Sarah; Cruaud, Corinne; Alberti, Adriana; Duarte, Carlos M.; Gasol, Josep M.; Vaqué, Dolors; Bork, Peer; Acinas, Silvia G.; Wincker, Patrick; Sullivan, Matthew B. (2016). "Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses". Nature. 537 (7622): 689–693. Bibcode:2016Natur.537..689.. doi:10.1038/nature19366. hdl:10754/619775. PMID 27654921. S2CID 54182070.
- ^ Gregory, Ann C.; Zayed, Ahmed A.; Conceição-Neto, Nádia; Temperton, Ben; Bolduc, Ben; Alberti, Adriana; Ardyna, Mathieu; Arkhipova, Ksenia; Carmichael, Margaux; Cruaud, Corinne; Dimier, Céline; Domínguez-Huerta, Guillermo; Ferland, Joannie; Kandels, Stefanie; Liu, Yunxiao; Marec, Claudie; Pesant, Stéphane; Picheral, Marc; Pisarev, Sergey; Poulain, Julie; Tremblay, Jean-Éric; Vik, Dean; Babin, Marcel; Bowler, Chris; Culley, Alexander I.; De Vargas, Colomban; Dutilh, Bas E.; Iudicone, Daniele; Karp-Boss, Lee; et al. (2019). "Marine DNA Viral Macro- and Microdiversity from Pole to Pole". Cell. 177 (5): 1109–1123.e14. doi:10.1016/j.cell.2019.03.040. PMC 6525058. PMID 31031001.
- ^ Nelson, David M.; Tréguer, Paul; Brzezinski, Mark A.; Leynaert, Aude; Quéguiner, Bernard (1995). "Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation". Global Biogeochemical Cycles. 9 (3): 359–372. Bibcode:1995GBioC...9..359N. doi:10.1029/95GB01070.
- ^ Malviya, Shruti; Scalco, Eleonora; Audic, Stéphane; Vincent, Flora; Veluchamy, Alaguraj; Poulain, Julie; Wincker, Patrick; Iudicone, Daniele; De Vargas, Colomban; Bittner, Lucie; Zingone, Adriana; Bowler, Chris (2016). "Insights into global diatom distribution and diversity in the world's ocean". Proceedings of the National Academy of Sciences. 113 (11): E1516–E1525. Bibcode:2016PNAS..113E1516M. doi:10.1073/pnas.1509523113. PMC 4801293. PMID 26929361.
- ^ a b Tréguer, Paul; Bowler, Chris; Moriceau, Brivaela; Dutkiewicz, Stephanie; Gehlen, Marion; Aumont, Olivier; Bittner, Lucie; Dugdale, Richard; Finkel, Zoe; Iudicone, Daniele; Jahn, Oliver; Guidi, Lionel; Lasbleiz, Marine; Leblanc, Karine; Levy, Marina; Pondaven, Philippe (2018). "Influence of diatom diversity on the ocean biological carbon pump". Nature Geoscience. 11 (1): 27–37. Bibcode:2018NatGe..11...27T. doi:10.1038/s41561-017-0028-x. S2CID 134885922.
- ^ Mahadevan, Amala; d'Asaro, Eric; Lee, Craig; Perry, Mary Jane (2012). "Eddy-Driven Stratification Initiates North Atlantic Spring Phytoplankton Blooms". Science. 337 (6090): 54–58. Bibcode:2012Sci...337...54M. doi:10.1126/science.1218740. PMID 22767922. S2CID 42312402.
- ^ Boyd, Philip W.; Claustre, Hervé; Levy, Marina; Siegel, David A.; Weber, Thomas (2019). "Multi-faceted particle pumps drive carbon sequestration in the ocean" (PDF). Nature. 568 (7752): 327–335. Bibcode:2019Natur.568..327B. doi:10.1038/s41586-019-1098-2. PMID 30996317. S2CID 119513489.
- ^ Behrenfeld, Michael J.; Doney, Scott C.; Lima, Ivan; Boss, Emmanuel S.; Siegel, David A. (2013). "Annual cycles of ecological disturbance and recovery underlying the subarctic Atlantic spring plankton bloom". Global Biogeochemical Cycles. 27 (2): 526–540. Bibcode:2013GBioC..27..526B. doi:10.1002/gbc.20050. S2CID 29575022.
- ^ Field, C. B.; Behrenfeld, M. J.; Randerson, J. T.; Falkowski, P. (1998). "Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components". Science. 281 (5374): 237–240. Bibcode:1998Sci...281..237F. doi:10.1126/science.281.5374.237. PMID 9657713.
- ^ Behrenfeld, M. J.; Randerson, J. T.; McClain, C. R.; Feldman, G. C.; Los, S. O.; Tucker, C. J.; Falkowski, P. G.; Field, C. B.; Frouin, R.; Esaias, W. E.; Kolber, D. D.; Pollack, N. H. (2001). "Biospheric Primary Production During an ENSO Transition". Science. 291 (5513): 2594–2597. Bibcode:2001Sci...291.2594B. doi:10.1126/science.1055071. PMID 11283369. S2CID 38043167.
- ^ Boetius, A.; Albrecht, S.; Bakker, K.; Bienhold, C.; Felden, J.; Fernandez-Mendez, M.; Hendricks, S.; Katlein, C.; Lalande, C.; Krumpen, T.; Nicolaus, M.; Peeken, I.; Rabe, B.; Rogacheva, A.; Rybakova, E.; Somavilla, R.; Wenzhofer, F.; RV Polarstern ARK27-3-Shipboard Science Party (2013). "Export of Algal Biomass from the Melting Arctic Sea Ice". Science. 339 (6126): 1430–1432. Bibcode:2013Sci...339.1430B. doi:10.1126/science.1231346. PMID 23413190. S2CID 206545871.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ^ Pachiadaki, Maria G.; Sintes, Eva; Bergauer, Kristin; Brown, Julia M.; Record, Nicholas R.; Swan, Brandon K.; Mathyer, Mary Elizabeth; Hallam, Steven J.; Lopez-Garcia, Purificacion; Takaki, Yoshihiro; Nunoura, Takuro; Woyke, Tanja; Herndl, Gerhard J.; Stepanauskas, Ramunas (2017). "Major role of nitrite-oxidizing bacteria in dark ocean carbon fixation". Science. 358 (6366): 1046–1051. Bibcode:2017Sci...358.1046P. doi:10.1126/science.aan8260. PMID 29170234. S2CID 206661557.
- ^ Grzymski, Joseph J.; Riesenfeld, Christian S.; Williams, Timothy J.; Dussaq, Alex M.; Ducklow, Hugh; Erickson, Matthew; Cavicchioli, Ricardo; Murray, Alison E. (2012). "A metagenomic assessment of winter and summer bacterioplankton from Antarctica Peninsula coastal surface waters". The ISME Journal. 6 (10): 1901–1915. doi:10.1038/ismej.2012.31. PMC 3446801. PMID 22534611.
- ^ Boetius, Antje; Wenzhöfer, Frank (2013). "Seafloor oxygen consumption fuelled by methane from cold seeps". Nature Geoscience. 6 (9): 725–734. Bibcode:2013NatGe...6..725B. doi:10.1038/ngeo1926.
- ^ Danovaro, Roberto; Corinaldesi, Cinzia; Dell'Anno, Antonio; Fuhrman, Jed A.; Middelburg, Jack J.; Noble, Rachel T.; Suttle, Curtis A. (2011). "Marine viruses and global climate change". FEMS Microbiology Reviews. 35 (6): 993–1034. doi:10.1111/j.1574-6976.2010.00258.x. PMID 21204862.
- ^ a b Schmidtko, Sunke; Stramma, Lothar; Visbeck, Martin (2017). "Decline in global oceanic oxygen content during the past five decades". Nature. 542 (7641): 335–339. Bibcode:2017Natur.542..335S. doi:10.1038/nature21399. PMID 28202958. S2CID 4404195.
- ^ a b Breitburg, Denise; Levin, Lisa A.; Oschlies, Andreas; Grégoire, Marilaure; Chavez, Francisco P.; Conley, Daniel J.; Garçon, Véronique; Gilbert, Denis; Gutiérrez, Dimitri; Isensee, Kirsten; Jacinto, Gil S.; Limburg, Karin E.; Montes, Ivonne; Naqvi, S. W. A.; Pitcher, Grant C.; Rabalais, Nancy N.; Roman, Michael R.; Rose, Kenneth A.; Seibel, Brad A.; Telszewski, Maciej; Yasuhara, Moriaki; Zhang, Jing (2018). "Declining oxygen in the global ocean and coastal waters". Science. 359 (6371): eaam7240. Bibcode:2018Sci...359M7240B. doi:10.1126/science.aam7240. PMID 29301986. S2CID 206657115.
- ^ a b Bertagnolli, Anthony D.; Stewart, Frank J. (2018). "Microbial niches in marine oxygen minimum zones". Nature Reviews Microbiology. 16 (12): 723–729. doi:10.1038/s41579-018-0087-z. PMID 30250271. S2CID 52811177.
- ^ a b Danovaro, Roberto; Molari, Massimiliano; Corinaldesi, Cinzia; Dell'Anno, Antonio (2016). "Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems". Science Advances. 2 (4): e1500961. Bibcode:2016SciA....2E0961D. doi:10.1126/sciadv.1500961. PMC 4928989. PMID 27386507.
- ^ Bienhold, Christina; Zinger, Lucie; Boetius, Antje; Ramette, Alban (2016). "Diversity and Biogeography of Bathyal and Abyssal Seafloor Bacteria". PLOS ONE. 11 (1): e0148016. Bibcode:2016PLoSO..1148016B. doi:10.1371/journal.pone.0148016. PMC 4731391. PMID 26814838. S2CID 13383023.
- ^ Rosenfeld, Daniel; Zhu, Yannian; Wang, Minghuai; Zheng, Youtong; Goren, Tom; Yu, Shaocai (2019). "Aerosol-driven droplet concentrations dominate coverage and water of oceanic low-level clouds". Science. 363 (6427): eaav0566. doi:10.1126/science.aav0566. PMID 30655446. S2CID 58612273.
- ^ a b Charlson, Robert J.; Lovelock, James E.; Andreae, Meinrat O.; Warren, Stephen G. (1987). "Oceanic phytoplankton, atmospheric sulphur, cloud albedo and climate". Nature. 326 (6114): 655–661. Bibcode:1987Natur.326..655C. doi:10.1038/326655a0. S2CID 4321239.
- ^ a b Gantt, B.; Meskhidze, N. (2013). "The physical and chemical characteristics of marine primary organic aerosol: A review". Atmospheric Chemistry and Physics. 13 (8): 3979–3996. Bibcode:2013ACP....13.3979G. doi:10.5194/acp-13-3979-2013.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Meskhidze, N.; Nenes, A. (2006). "Phytoplankton and Cloudiness in the Southern Ocean". Science. 314 (5804): 1419–1423. Bibcode:2006Sci...314.1419M. doi:10.1126/science.1131779. PMID 17082422. S2CID 36030601.
- ^ Andreae, M.O.; Rosenfeld, D. (2008). "Aerosol–cloud–precipitation interactions. Part 1. The nature and sources of cloud-active aerosols". Earth-Science Reviews. 89 (1–2): 13–41. Bibcode:2008ESRv...89...13A. doi:10.1016/j.earscirev.2008.03.001.
- ^ Moore, R. H.; Karydis, V. A.; Capps, S. L.; Lathem, T. L.; Nenes, A. (2013). "Droplet number uncertainties associated with CCN: An assessment using observations and a global model adjoint". Atmospheric Chemistry and Physics. 13 (8): 4235–4251. Bibcode:2013ACP....13.4235M. doi:10.5194/acp-13-4235-2013.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Sanchez, Kevin J.; Chen, Chia-Li; Russell, Lynn M.; Betha, Raghu; Liu, Jun; Price, Derek J.; Massoli, Paola; Ziemba, Luke D.; Crosbie, Ewan C.; Moore, Richard H.; Müller, Markus; Schiller, Sven A.; Wisthaler, Armin; Lee, Alex K. Y.; Quinn, Patricia K.; Bates, Timothy S.; Porter, Jack; Bell, Thomas G.; Saltzman, Eric S.; Vaillancourt, Robert D.; Behrenfeld, Mike J. (2018). "Substantial Seasonal Contribution of Observed Biogenic Sulfate Particles to Cloud Condensation Nuclei". Scientific Reports. 8 (1): 3235. Bibcode:2018NatSR...8.3235S. doi:10.1038/s41598-018-21590-9. PMC 5818515. PMID 29459666.
- ^ a b Atwood, Trisha B.; Connolly, Rod M.; Ritchie, Euan G.; Lovelock, Catherine E.; Heithaus, Michael R.; Hays, Graeme C.; Fourqurean, James W.; MacReadie, Peter I. (2015). "Predators help protect carbon stocks in blue carbon ecosystems". Nature Climate Change. 5 (12): 1038–1045. Bibcode:2015NatCC...5.1038A. doi:10.1038/nclimate2763.
- ^ Myers, Ransom A.; Worm, Boris (2003). "Rapid worldwide depletion of predatory fish communities". Nature. 423 (6937): 280–283. Bibcode:2003Natur.423..280M. doi:10.1038/nature01610. PMID 12748640. S2CID 2392394.
- ^ Duarte, Carlos M.; Losada, Iñigo J.; Hendriks, Iris E.; Mazarrasa, Inés; Marbà, Núria (2013). "The role of coastal plant communities for climate change mitigation and adaptation". Nature Climate Change. 3 (11): 961–968. Bibcode:2013NatCC...3..961D. doi:10.1038/nclimate1970.
Protist shell
[edit]"The amoebae also are extremely diverse. Amoebae are defined based on pseudopodia type: those with thin, or filose, pseudopods, which may be reinforced by stiff microtubule proteins, are classified in the supergroup Rhizaria (e.g., foraminiferans and radiolarians), whereas those with lobose pseudopods, which are blunt and are not reinforced, are classified in the supergroup Amoebozoa. Both groups of amoebae can be “naked” or housed inside a shell, or test, composed of organic or inorganic materials.The naked amoebae are the simplest of the amoebae. They have no defined shape and extend one or many lobose pseudopodia. Many of these lobose amoebae, including those in the genera Mastigamoeba and Mastigella, also possess flagella in the vegetative (resting) phase. At the opposite extreme are the complex foraminiferans, which live inside multichambered calcareous shells up to several millimetres in diameter. The filose pseudopodia of foraminiferans are known as reticulopodia and extend from the aperture of the largest chamber of the shell, forming a complicated, sticky branching network. Rhizarian amoebae that are known commonly as radiolarians form shells from silica or strontium sulfate; in some the shell has so many holes that the structure resembles a sponge. The polyphyletic heliozoans, or sun protozoans, have radiating pseudopodia (axopodia) that extend like spokes from the central body; microtubules support an outer layer of cytoplasm. Many heliozoans are members of Rhizaria; however, some are placed in Chromalveolata."[1]
Dinoflagellates
[edit]Dinoflagellates are part of the algae group, and form a phylum of unicellular flagellates with about 2,000 marine species.[2] The name comes from the Greek "dinos" meaning whirling and the Latin "flagellum" meaning a whip or lash. This refers to the two whip-like attachments (flagella) used for forward movement. Most dinoflagellates are protected with red-brown, cellulose armour. Like other phytoplankton, dinoflagellates are r-strategists which under right conditions can bloom and create red tides. Excavates may be the most basal flagellate lineage.[3]
By trophic orientation dinoflagellates cannot be uniformly categorized. Some dinoflagellates are known to be photosynthetic, but a large fraction of these are in fact mixotrophic, combining photosynthesis with ingestion of prey (phagotrophy).[4] Some species are endosymbionts of marine animals and other protists, and play an important part in the biology of coral reefs. Others predate other protozoa, and a few forms are parasitic. Many dinoflagellates are mixotrophic and could also be classified as phytoplankton.
-
Gyrodinium, one of the few naked dinoflagellates which lack armour
-
The dinoflagellate Dinophysis acuta
-
Tripos muelleri is recognisable by its U-shaped horns
Amoeba
[edit]-
Naked amoeba showing food vacuoles and ingested diatom
-
Shell or test of a testate amoeba, Arcella sp.
-
Xenogenic testate amoeba covered in diatoms (from Penard's Amoeba Collection)
Ciliates
[edit]-
Tintinnopsis campanula
-
Holophyra ovum
-
Mesodinium rubrum produce deep red blooms using enslaved chloroplasts from their algal prey [5]
-
Several taxa of ciliates interacting
-
Blepharisma americanum swimming in a drop of pond water with other microorganisms
- Mixoplankton
-
Tintinnid ciliate Favella
-
Euglena mutabilis, a photosynthetic flagellate
-
Zoochlorellae (green) living inside the ciliate Stichotricha secunda
Other taxa
[edit]Microfossils
[edit]
Sediments at the bottom of the ocean have two main origins, terrigenous and biogenous.
Terrigenous sediments account for about 45% of the total marine sediment, and originate in the erosion of rocks on land, transported by rivers and land runoff, windborne dust, volcanoes, or grinding by glaciers.
Biogenous sediments account for the other 55% of the total sediment, and originate in the skeletal remains of marine protists (single-celled plankton and benthos microorganisms). Much smaller amounts of precipitated minerals and meteoric dust can also be present. Ooze, in the context of a marine sediment, does not refer to the consistency of the sediment but to its biological origin. The term ooze was originally used by John Murray, the "father of modern oceanography", who proposed the term radiolarian ooze for the silica deposits of radiolarian shells brought to the surface during the Challenger Expedition.[9] A biogenic ooze is a pelagic sediment containing at least 30 percent from the skeletal remains of marine organisms.

Main types of biogenic ooze
| ||||||||
|---|---|---|---|---|---|---|---|---|
| type | mineral forms |
protist involved |
name of skeleton | typical size (mm) |
||||
| Siliceous ooze | SiO2 silica quartz glass opal chert |
diatom | 
|
frustule | 0.002 to 0.2[10] | 
|
diatom microfossil from 40 million years ago | |
| radiolarian | 
|
test or shell | 0.1 to 0.2 | 
|
elaborate silica shell of a radiolarian | |||
| Calcareous ooze | CaCO3 calcite aragonite limestone marble chalk |
foraminiferan | 
|
test or shell | under 1 | 
|
Calcified test of a planktic foraminiferan. There are about 10,000 living species of foraminiferans[11] | |
| coccolithophore | 
|
coccoliths | under 0.1[12] | 
|
Coccolithophores are the largest global source of biogenic calcium carbonate, and significantly contribute to the global carbon cycle.[13] They are the main constituent of chalk deposits such as the white cliffs of Dover. | |||
-
Diatomaceous earth is a soft, siliceous, sedimentary rock made up of microfossils in the form of the frustules (shells) of single cell diatoms (click to magnify)
-
Illustration of a Globigerina ooze
-
Shells (tests), usually made of calcium carbonate, from a foraminiferal ooze on the deep ocean floor

-
Opal can contain protist microfossils of diatoms, radiolarians, silicoflagellates and ebridians [15]
-
Marble can contain protist microfossils of foraminiferans, coccolithophores, calcareous nannoplankton and algae, ostracodes, pteropods, calpionellids and bryozoa [15]


Within each colored area, the type of material shown is what dominates, although other materials are also likely to be present.
For further information, see here
References
[edit]- ^ Amoebae and pseudopodia
- ^ Gómez F (2012). "A checklist and classification of living dinoflagellates (Dinoflagellata, Alveolata)" (PDF). CICIMAR Océanides. 27 (1): 65–140. doi:10.37543/oceanides.v27i1.111. Archived from the original (PDF) on 2013-11-27.
- ^ Cite error: The named reference
Dawson2013was invoked but never defined (see the help page). - ^ Stoecker DK (1999). "Mixotrophy among Dinoflagellates". The Journal of Eukaryotic Microbiology. 46 (4): 397–401. doi:10.1111/j.1550-7408.1999.tb04619.x. S2CID 83885629.
- ^ Dierssen, Heidi; McManus, George B.; Chlus, Adam; Qiu, Dajun; Gao, Bo-Cai; Lin, Senjie (2015). "Space station image captures a red tide ciliate bloom at high spectral and spatial resolution". Proceedings of the National Academy of Sciences. 112 (48): 14783–14787. Bibcode:2015PNAS..11214783D. doi:10.1073/pnas.1512538112. PMC 4672822. PMID 26627232.
- ^ Booth, B.C. and Marchant, H.J. (1987) "Parmales, a new order of marine chrysophytes, with descriptions of three new genera and seven new species". Journal of Phycology, 23: 245–260. doi:10.1111/j.1529-8817.1987.tb04132.x.
- ^ Kuwata, A., Yamada, K., Ichinomiya, M., Yoshikawa, S., Tragin, M., Vaulot, D. and Lopes dos Santos, A. (2018) "Bolidophyceae, a sister picoplanktonic group of diatoms – a review". Frontiers in Marine Science, 5: 370. doi:10.3389/fmars.2018.00370.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Yasuhara, Moriaki; Huang, Huai-Hsuan; Hull, Pincelli; Rillo, Marina; Condamine, Fabien; Tittensor, Derek; Kučera, Michal; Costello, Mark; Finnegan, Seth; o'Dea, Aaron; Hong, Yuanyuan; Bonebrake, Timothy; McKenzie, Ryan; Doi, Hideyuki; Wei, Chih-Lin; Kubota, Yasuhiro; Saupe, Erin (2020). "Time Machine Biology: Cross-Timescale Integration of Ecology, Evolution, and Oceanography". Oceanography. 33 (2). doi:10.5670/oceanog.2020.225. S2CID 225850315.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Thomson, Charles Wyville (2014) Voyage of the Challenger : The Atlantic Cambridge University Press, page235. ISBN 9781108074759.
- ^ Grethe R. Hasle; Erik E. Syvertsen; Karen A. Steidinger; Karl Tangen (1996-01-25). "Marine Diatoms". In Carmelo R. Tomas (ed.). Identifying Marine Diatoms and Dinoflagellates. Academic Press. pp. 5–385. ISBN 978-0-08-053441-1. Retrieved 2013-11-13.
- ^ Ald, S.M.; et al. (2007). "Diversity, Nomenclature, and Taxonomy of Protists" (PDF). Syst. Biol. 56 (4): 684–689. doi:10.1080/10635150701494127. PMID 17661235. Archived from the original (PDF) on 31 March 2011. Retrieved 11 October 2019.
- ^ Moheimani, N.R.; Webb, J.P.; Borowitzka, M.A. (2012), "Bioremediation and other potential applications of coccolithophorid algae: A review. . Bioremediation and other potential applications of coccolithophorid algae: A review", Algal Research, 1 (2): 120–133, doi:10.1016/j.algal.2012.06.002
- ^ Taylor, A.R.; Chrachri, A.; Wheeler, G.; Goddard, H.; Brownlee, C. (2011). "A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores". PLOS Biology. 9 (6): e1001085. doi:10.1371/journal.pbio.1001085. PMC 3119654. PMID 21713028.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Wierer, U.; Arrighi, S.; Bertola, S.; Kaufmann, G.; Baumgarten, B.; Pedrotti, A.; Pernter, P.; Pelegrin, J. (2018). "The Iceman's lithic toolkit: Raw material, technology, typology and use". PLOS ONE. 13 (6): e0198292. Bibcode:2018PLoSO..1398292W. doi:10.1371/journal.pone.0198292. PMC 6010222. PMID 29924811.
- ^ a b Haq B.U. and Boersma A. (Eds.) (1998) Introduction to Marine Micropaleontology Elsevier. ISBN 9780080534961
Further references
[edit]- Xu, K., Hutchins, D. and Gao, K. (2018) "Coccolith arrangement follows Eulerian mathematics in the coccolithophore Emiliania huxleyi". PeerJ, 6: e4608. doi:10.1126/science.aaa7378.
Marine coastal ecosystems
[edit]
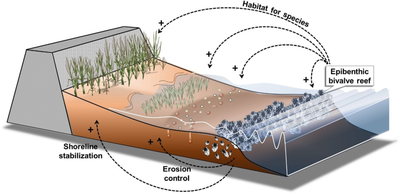
- references
Valuation of marine ecosystems
[edit]
- references
- ^ Cavanagh, Rachel D.; Broszeit, Stefanie; Pilling, Graham M.; Grant, Susie M.; Murphy, Eugene J.; Austen, Melanie C. (2016). "Valuing biodiversity and ecosystem services: A useful way to manage and conserve marine resources?". Proceedings of the Royal Society B: Biological Sciences. 283 (1844). doi:10.1098/rspb.2016.1635. PMC 5204147. PMID 27928037.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Bacteria morphology
[edit]
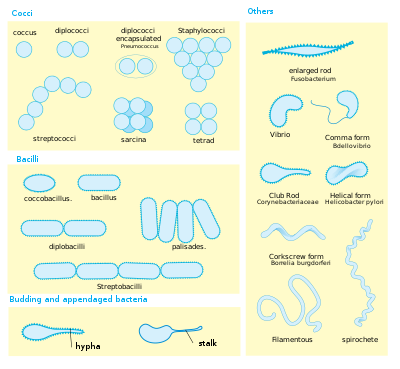
FROM: Coccus...
A coccus (plural cocci) is any bacterium or archaeon that has a spherical, ovoid, or generally round shape.[2][3] Bacteria are categorized based on their shapes into three classes: cocci (spherical-shaped), bacillus (rod-shaped) and spirochetes (spiral-shaped) cells.[3] Coccus refers to the shape of the bacteria, and can contain multiple genera, such as staphylococci or streptococci. Cocci can grow in pairs, chains, or clusters, depending on their orientation and attachment during cell division. Contrast to many bacilli-shaped bacteria, most cocci bacteria do not have flagella and are non-motile.[4]
FROM Bacillus (shape)...
A bacillus (plural bacilli), or bacilliform bacterium, is a rod-shaped bacterium or archaeon. Bacilli are found in many different taxonomic groups of bacteria. However, the name Bacillus, capitalized and italicized, refers to a specific genus of bacteria. The name Bacilli, capitalized but not italicized, can also refer to a less specific taxonomic group of bacteria that includes two orders, one of which contains the genus Bacillus. When the word is formatted with lowercase and not italicized, 'bacillus', it will most likely be referring to shape and not to the genus at all. Bacilliform bacteria are also often simply called rods when the bacteriologic context is clear.[5]
FROM Spiral bacteria...
Spiral bacteria, bacteria of spiral (helical) shape, form the third major morphological category of prokaryotes along with the rod-shaped bacilli and round cocci.[6][7] Spiral bacteria can be subclassified by the number of twists per cell, cell thickness, cell flexibility, and motility. The two types of spiral cells are spirillum and spirochete, with spirillum being rigid with external flagella, and spirochetes being flexible with internal flagella.[8]
- references
- ^ Schulz, Kestin; Smit, Mariya W.; Herfort, Lydie; Simon, Holly M. (2018). "The Unseen World in the River". Frontiers for Young Minds. 6. doi:10.3389/frym.2018.00004. S2CID 3344238.
- ^ Pommerville, J.C. (2013). Fundamentals of Microbiology (10th ed.). Sudbury, MA: Jones & Bartlett. p. 106. ISBN 9781449647964.
- ^ a b Ryan, Kenneth James (4 January 2018). Sherris medical microbiology (7th ed.). New York: McGraw-Hill Education. ISBN 9781259859809. OCLC 983825627.
- ^ Levinson, Warren; Joyce, Elizabeth A.; Nussbaum, Jesse; Schwartz, Brian S.; Chin-Hong, Peter (10 May 2018). Review of medical microbiology & immunology: a guide to clinical infectious diseases (15th ed.). New York: McGraw-Hill Education. ISBN 9781259644498. OCLC 1032261353.
- ^ "The Size, Shape, And Arrangement Of Bacterial Cells". Midlands Technical College. Archived from the original on 9 August 2016. Retrieved 8 August 2016.
- ^ Csuros, Maria; Csuros, Csaba (1999). Microbiological Examination of Water and Wastewater. Boca Raton, Florida: CRC Press. pp. 16–17. ISBN 9781566701792.
- ^ Young, Kevin D. (September 2006). "The Selective Value of Bacterial Shape". Microbiology and Molecular Biology Reviews. 70 (3): 660–703. doi:10.1128/MMBR.00001-06. PMC 1594593. PMID 16959965.
- ^ Taro, Kathleen (2007). Foundations in Microbiology (6th International ed.). McGraw-Hill. pp. 108–109. ISBN 978-0071262323. Retrieved 11 September 2017.
Marine plankton
[edit]Freshwater fungi
[edit]NEEDS PARAPHRASING...
"Molecular diversity of microbial eukaryotes in aquatic ecosystems is far less investigated than their prokaryotic counterparts. This is even more striking for particular groups such as fungi that attract very little interest. This bias partly results from their supposed low abundances (e.g., ∼1% of total marine eukaryotes (Massana & Pedros-Alio, 2008)) that suggests fungi have little ecological importance in aquatic ecosystems. However, rare organisms can play crucial roles in ecosystem functioning but more importantly recent studies have revealed much larger proportions of fungi than previously observed, as well as high taxonomic richness in different marine (Le Calvez et al., 2009; Gao, Johnson & Wang, 2010; Orsi, Biddle & Edgcomb, 2013; Richards et al., 2012; Lepère et al., 2015) and freshwaters environments (Monchy et al., 2011; Ishii, Ishida & Kagami, 2015; Duarte et al., 2015; Lepère et al., 2016). The extent of fungal biodiversity is therefore likely underestimated (Scheffers et al., 2012) though diversity estimates based on molecular data suggest that it can range between 0.5 and 10 million species (Hawksworth, 2001; O’Brien et al., 2005; Mora et al., 2011; Bass & Richards, 2011; Blackwell, 2011)."[1]
NEEDS PARAPHRASING...
"Despite this putative rich biodiversity, functional roles of aquatic fungi, for which only 3,000–4,000 species have been recorded, remain poorly characterized (Pautasso, 2013; Rambold, Stadler & Begerow, 2013). They are mainly known as decomposers of leaves in rivers, mangroves and wetlands (Seena, Wynberg & Bärlocher, 2008; Gulis, Kuehn & Suberkropp, 2009) and as parasites of phytoplankton and zooplankton in lake ecosystems (Jobard, Rasconi & Sime-Ngando, 2010). A decade ago, fungi were divided into four main phyla: Basidiomycota, Ascomycota, Zygomycota and Chytridiomycota (James et al., 2006). However, phylogeny of fungi is still unresolved and several phyla, classes and orders of basal fungi have been determined, since. For example, Corradi (2015) highlighted that Cryptomycota forms a new phylum in which we can find Microsporidia, Aphelids and Rozellids. The vast majority of this phylum is characterized by environmental sequences and gathered under the term “dark matter fungi” (Grossart et al., 2015). These fungi are mostly zoosporic and “old” in term of evolution since they diverged from the remaining fungi 710–1,060 million years ago (Lücking et al., 2009). These basal fungi are distant from cultured and described fungi. Aquatic environments are thus likely to host a high number of uncharacterized groups (Grossart & Rojas-Jimenez, 2016)."[1]
Bacterial and fungal communities (in glacier-fed streams)
[edit]

NEEDS PARAPHRASING...
"Bacterial and fungal communities in biofilms are important components in driving biogeochemical processes in stream ecosystems. Previous studies have well documented the patterns of bacterial alpha diversity in stream biofilms in glacier-fed streams, where, however, beta diversity of the microbial communities has received much less attention especially considering both bacterial and fungal communities. A focus on beta diversity can provide insights into the mechanisms driving community changes associated to large environmental fluctuations and disturbances, such as in glacier-fed streams. Moreover, modularity of co-occurrence networks can reveal more ecological and evolutionary properties of microbial communities beyond taxonomic groups."[2]
NEEDS PARAPHRASING...
"Glaciers cover approximately 10% of the land surface on the Earth (Milner et al., 2017) and are important components of the hydrological cycle providing vital water resources (Barnett, Adam & Lettenmaier, 2005; Gardner et al., 2013; Zemp et al., 2015). However, glaciers are shrinking rapidly across the world due to accelerating global warming (Immerzeel, Van Beek & Bierkens, 2010; Sorg et al., 2012; Marzeion et al., 2014), and most of them are expected to disappear by 2050 (Zemp et al., 2006; IPCC, 2014). As a prominent component of the glacier forefront, glacier-fed streams have a highly heterogeneous environment due to longitudinal alterations of landcover, river hydrology and morphology, sediment transport, and biogeochemical processes (Hood & Scott, 2008; Laghari, 2013; Hotaling, Hood & Hamilton, 2017; Milner et al., 2017). For example, from glacier terminus to downstream, terrestrial vegetation increases (Zhang et al., 2013; Raynolds et al., 2015), stream channel lengthens (Milner, Brown & Hannah, 2009; Robinson, Thompson & Freestone, 2014), and water source compositions changes (Brown, Hannah & Milner, 2003)."[2]

- Biofilm
"Stated simply, biofilms are microorganisms attached to a solid surface. Biofilms pervade virtually all environments, often dominating the microbial activity distributed between the individual planktonic and aggregated habitats"
NEEDS PARAPHRASING...
"Biofilms are hot spots of microbial diversity and activity in stream ecosystems (Geesey et al., 1978; Battin et al., 2016). Within stream biofilms, bacteria, fungi, and algae are the major components driving the bulk of metabolism and biogeochemical processes (Brittain & Milner, 2001; Battin et al., 2003; Von Schiller et al., 2007). The changing environment presents significant challenges for glacier-fed stream ecosystems. Previous studies have revealed that factors associated with glacier shrinkage have significant influences on the composition, diversity, and functional potential of bacterial communities in stream biofilms (Wilhelm et al., 2013, 2014; Ren, Gao & Elser, 2017; Ren et al., 2017). However, fungal communities in glacial systems are rarely studied (Edwards, 2015; Anesio et al., 2017). With the decrease in elevation, glacier coverage, and glacier source contribution to streamflow, as well as increase in distance to glacier terminus, bacterial communities showed increased alpha diversity as well as distinct taxonomic and functional compositions (Wilhelm et al., 2013, 2014; Ren, Gao & Elser, 2017; Ren et al., 2017)."[2]
FROM: Biofilm...
Biofilms can be found on rocks and pebbles at the bottoms of most streams or rivers and often form on the surfaces of stagnant pools of water. Biofilms are important components of food chains in rivers and streams and are grazed by the aquatic invertebrates upon which many fish feed.
- More biofilm
NEEDS PARAPHRASING...
"Streams and rivers form dense networks, shape the Earth’s surface and, in their sediments, provide an immensely large surface area for microbial growth. Biofilms dominate microbial life in streams and rivers, drive crucial ecosystem processes and contribute substantially to global biogeochemical fluxes. In turn, water flow and related deliveries of nutrients and organic matter to biofilms constitute major constraints on microbial life."[3]
NEEDS PARAPHRASING...
"The perception that most microorganisms live as complex communities that are attached to surfaces has profoundly changed microbiology over the past decades. Most, if not all, bacteria can form biofilms, which are communities of cells embedded in a porous extracellular matrix. Dental plaque, the microorganisms on catheters and implants that cause persistent infections and the fouling of ship hulls and pipework are all examples of biofilms with important implications for public health and industrial processes. Most contemporary biofilm research rests on the discovery made more than 35 years ago by Maurice Lock, Gill Geesey and Bill Costerton: bacteria attached to surfaces dominate microbial life in streams1–3. These microbiologists pioneered research into stream biofilms, also termed periphyton or epilithon, and described them as complex aggregates of bacteria, algae, protozoa, fungi and meiobenthos. The early study of stream biofilms also highlighted the relevance of interactions between microbial phototrophs and heterotrophs for energy fluxes and the role of the biofilm matrix as the site of extracellular enzyme activity and adsorption of dissolved organic matter (DOM)3,4."[3]
NEEDS PARAPHRASING...
"Since these early days, the study of the ecology and biogeochemistry of stream biofilms has slowly developed in the wake of thriving research on bacterial biofilms — often comprising only a single strain, of interest to medical microbiology, rather than the polymicrobial communities found in stream biofilms — and on the microbial ecology of marine and lake planktonic communities5. Unlike bacterial biofilms grown in the laboratory, biofilms in
streams are continuously exposed to a diverse inoculum that includes bacteria, archaea, algae, fungi, protozoa and even metazoa. These diverse biological ‘building blocks’, when combined with the dynamic flow of streamwater, generate biofilms with inherently complex and varying physical structures that have implications for microbial functioning and ecosystem processes6. In streams, biofilms are key sites of enzymatic activity7, including organic matter cycling, ecosystem respiration and primary production and, as such, form the basis of the food web."[3]
NEEDS PARAPHRASING...
"Why should we study the ecology and biogeochemistry of stream biofilms? Streams sculpt the continental surface, forming dense and conspicuous channel networks that can be thought of as ecological arteries that perfuse the landscape. Streams are connected to their catchments through various surface and subsurface flow paths and notably through the hyporheic zone in the streambed at the interface between groundwater and streamwater8. Microbial cells, solutes and particles enter streams through these flow paths and, en route to downstream ecosystems and ultimately to the oceans, they may interact with the biofilms that colonize the large surface area provided by the streambed as a ‘microbial skin’ (BOX 1). As a result, the streambed and its biofilm microbiome contribute to biogeochemical fluxes8. Indeed, stream biofilms are now recognized as substantial contributors to global carbon fluxes by degrading organic matter and ultimately emitting an unexpectedly large amount of carbon dioxide into the atmosphere9,10. Microorganisms in streams are also major components of the nitrogen cycle as they denitrify nitrate that they receive from catchment and emit the resulting nitrous oxide or nitrogen gas into the atmosphere11,12. Furthermore, stream biofilms can be viewed as a crucial component of the catchment microbiome that also includes the microbial communities of the phyllosphere13 and soil14. The phyllosphere and the soil crust intercept water, microorganisms and solutes upon their entry into the catchment, whereas stream biofilms regulate the export of microorganisms and solutes from the catchment. Stream biofilms thus connect the land surface, groundwater, oceans and the atmosphere, and as such they are prominently positioned at the nexus of global biogeochemistry, biodiversity and climate change."[3]
- Beta diversity
NEEDS PARAPHRASING...
"Biodiversity is important for generating and stabilizing ecosystem structure and functions (Loreau et al., 2001; Tilman, Isbell & Cowles, 2014). The positive effects of local species richness (alpha diversity) on ecosystem functioning have been widely confirmed by a growing number of studies (Chapin et al., 2000; Cardinale et al., 2012; Duffy, Godwin & Cardinale, 2017). However, comparing to alpha diversity, beta diversity is an underexplored facet of biodiversity (Mori, Isbell & Seidl, 2018), which accumulates from compositional variations among local assemblages and provides insights into the mechanisms underlining biodiversity changes and their ecological consequences (Anderson et al., 2011; Socolar et al., 2016). For ecological communities suffering intensive environmental fluctuations and disturbances, focusing on beta diversity is especially important (Mori, Isbell & Seidl, 2018). In addition, microorganisms in many environments often coexist in a complex network with positive and negative interactions among members, playing pivotal roles in community assembly (Fuhrman, 2009; Barberán et al., 2012; Shi et al., 2016). These interactions may imply biologically or biochemically meaningful relationships between microorganisms (Weiss et al., 2016). Microbial co-occurrence networks can reveal how taxa potentially interact with each other, how diverse taxa structure networks, and how networks are compartmentalized into modules of closely associated taxa, as well as how microbial communities responded to environmental variations (Newman, 2006; Fuhrman, 2009; De Menezes et al., 2015; Banerjee et al., 2016). In addition, modularity (the tendency of a network to contain sub-clusters of nodes) is an important ecological feature in many biological systems, providing opportunities to identify highly connected taxa and integrate high dimension data into predicted ecological modules (De Menezes et al., 2015; Shi et al., 2016). A module is defined as a group of densely connected operational taxonomic units (OTUs), which have less links with OTUs belonging to other modules (Shi et al., 2016), forming a clustered network topology (Barberán et al., 2012). Modules can help to reveal more ecological and evolutionary properties (Thompson, 2005; Olesen et al., 2007), which are easily overlooked when communities are studied as a whole or in taxonomic groups (Porter, Onnela & Mucha, 2009; Bissett et al., 2013; De Menezes et al., 2015). The relationships between microbial modules and environmental variables can improve our understanding of the influences of environmental variation on microbial community assembly (Lindström & Langenheder, 2012; De Menezes et al., 2015; Toju et al., 2016). However, previous studies in glacier-fed streams have only focused on the whole communities or certain taxonomic groups of bacteria and fungi (Robinson & Jolidon, 2005; Milner, Brown & Hannah, 2009; Wilhelm et al., 2013; Ren, Gao & Elser, 2017). The network and modularity features of bacterial and fungal communities in glacier-fed streams are remaining one of our knowledge gaps. Integrating beta diversity and network modularity can provide novel insights into assembly mechanisms of microbial communities in glacier-fed streams."[2]
- Biodiversity across spatial scales
NEEDS PARAPHRASING...
"Stream biofilms are jungles of biodiversity, and the organisms that are typically found within them span across the entire tree of life. The development of next-generation sequencing methods has enabled a high-throughput profiling of these biofilms that has impressively demonstrated the full breadth and complexity of their microbial diversity (FIG. 1). Depending on light availability, eukaryotic algae (such as diatoms, green algae, chrysophytes, red algae and cryptophytes) and cyanobacteria, together with bacteria and to some extent also archaea,
can form biofilms in the benthic zone, whereas bacteria and archaea dominate in the deeper
sediments in which phototrophic life is limited. Fungi are probably also an important component of stream biofilms but remain poorly studied15,16. Ciliates, flagellates, nematodes and even young-instar insects (such as midges) are among the top consumers in stream biofilms17,18, and their grazing activity can change the physical structure19,20, community composition21 and carbon cycling of biofilms22. Furthermore, viruses have an important role in marine ecosystems23; for example, in bacterial biofilms24, bacterial viruses (or phages) can infect cells and regulate the dynamics and diversity of bacterial communities. However, little is known of the abundance and relevance of viruses in stream biofilms."[3]
- Marine biofilm
Freshwater prokaryotes
[edit]

Lake sediments
[edit]Lake sediments harbor diverse microbial communities that cycle carbon and nutrients while being constantly colonized and potentially buried by organic matter sinking from the water column.[5]
The continuous deposition of organic and inorganic particles to sediments is an important process in all aquatic ecosystems. Approximately one third of the terrestrial organic matter (OM) that enters freshwater is sequestered in sediments,[6] although the total amount of OM that reaches the sediments is much greater than the amount that is actually sequestered.[7] This is because microbial activity is responsible for the cycling of carbon, including methane emission.[8] In lake sediments, a proportion of newly settled OM is rapidly recycled and subsequently transformed into secondary compounds, resulting in a distinct uppermost sediment zone of high heterotrophic activity.[9][10] This is thought to lead to the structuring of microbial communities along environmental gradients that are much steeper than those in marine sediments, with narrower vertical sequences of electron acceptors.[11] The nature of this gradient influences the carbon, nitrogen, and sulfur cycles [12][13][14] and potentially affects the microbial community structure.[15][5]
In contrast to the wealth of studies on marine sediments (cf. the 65 studies of,[16] few studies have examined the vertical microbial community structure of freshwater sediments (e.g.,[17][18][19][20][21]). The community of sediment microbes was thought to be dominated by Bacteria, together with a smaller fraction of methanogenic Archaea.[22][23][24] This view has been challenged by the recent discovery of an abundance of non-methanogenic Archaea in marine sediments.[25][26] They are assumed to be adapted to low-energy environments, and at least one lineage seems to be specialized in inter alia amino acid turnover.[26] This discovery has led to a revised perception of microbial communities in marine sediments, where Archaea appear to be as abundant as Bacteria and increase in relative abundance with sediment depth.[16] Data on sediment Archaea in freshwater are scarce, and the causes of the significant variation observed among studies remain largely unknown.[19][27][28][29][5]
Prokaryotic activity, biomass, and cell numbers decrease with depth in many freshwater and marine sediments,[30][31] although other studies report relatively constant proportions of active cells with depth and find no accumulation of dead cells in deeper sediments.[32][33] Despite the continuous presence of vegetative cells and resting stages, recent studies of marine systems indicate that the majority of microbial cells in energy-deprived horizons consist of microbial necromass [34][35] and the proportion of living organisms decreases with the increasing age of the sediment [29]. The vertical, progressive transformation of OM and depletion of electron acceptors may eventually lead to an extremely low-energy environment in deeper sediment layers with very low growth rates similar to sub-seafloor sediments.[36][5]
NEEDS PARAPHRASING...
"Lakes and ponds are the final resting place for many of the Earth’s plants. Rivers collect much of the planet’s dead organic matter, transporting it to rest in calmer waters. But on a microscopic scale, lakes are anything but calm. An invisible metropolis of microbes feeds on these logs and leaves, producing greenhouse gases as a byproduct. As a result, lakes may be responsible for as much as a quarter of the carbon in the atmosphere – and rising. New research conducted with my colleagues in Cambridge, Germany and Canada suggests that emissions from freshwater lakes could double in the coming decades because of climate change. All known life on Earth is made of carbon. When plants and animals reach the end of their lives, microorganisms such as bacteria and fungi come to feast. They feed on the carbon-based remains of other organisms and their waste products — collectively known as organic matter."[37]
Comparison with marine prokaryotes
[edit]"Phylogenetic relationships of organisms within and across ecosystems can provide insight into the evolutionary history of lineages and how evolution might proceed into the future. Microorganisms in the water columns of freshwater and marine ecosystems provide a unique juxtaposition. On one hand, these habitats share common features of pelagic lifestyles like free-living and particle-associated niches,[38] potential for interactions with phytoplankton,[39] and opportunities for diverse photoheterotrophic organisms, including aerobic anoxygenic phototrophs [40] and rhodopsin-containing bacteria.[41][42] However, salinity preference is considered a complex trait involving many genes and complex cellular integration,[43][44] suggesting that transitions between high and low salinity are difficult from a genetic perspective. Consistent with this idea, microbial communities from saline environments are compositionally distinct from those inhabiting nonsaline environments.[45][46] Salinity-induced shifts in microbial beta diversity have been observed in studies of marine-to-freshwater gradients in many systems, including the Baltic Sea,[44][47] Columbia River Estuary system,[48] and Antarctic lakes that have become progressively less saline since becoming isolated from the sea.[49] These observations of ecosystem-specific diversity support the current paradigm that transitions between marine and freshwater ecosystems are infrequent, despite many ecological similarities.[50][51]

The abundances of phyla (classes for Proteobacteria), orders, and families between marine and freshwater samples were compared. At the phylum level, Alphaproteobacteria, Gammaproteobacteria, Euryarchaeota, and Marinimicrobia had significantly higher relative abundances in marine systems while Betaproteobacteria and Verrucomicrobia had higher relative abundances in freshwater systems.[51]

"Environmental sequence data provide support for a "salty divide" separating marine and freshwater microbial assemblages. From a phylogenetic perspective, each clade that contains both marine and freshwater representatives includes at least one transition where a common ancestor gave rise to a daughter lineage able to survive and proliferate in a new salinity environment.[51] Transitions that occurred recently are expected to result in highly similar molecular sequences recovered from marine and freshwater systems while transitions that occurred in the distant past are expected to yield habitat-specific diversification — clades that are only observed in one habitat type or the other — and a greater sequence divergence between marine and freshwater representatives.[51] Prior work using phylogenetic patterns concluded that transitions between marine and freshwater environments are infrequent and most transition events occurred a long time ago in evolutionary terms.[51][52] For example, Logares and colleagues [52] found that within the abundant alphaproteobacterial SAR11 group, freshwater representatives belonged exclusively to a single subclade, called LD12, implying a single salinity transition from a marine ancestor to this freshwater lineage. Besides LD12, there are a number of microbial lineages that appear to be unique to freshwater lakes,[53][54] suggesting that these lineages do not readily colonize other habitat types. Notably, for freshwater lineages that are found in multiple habitats, the secondary habitat is most often terrestrial, not marine,[54] consistent with the idea that marine-freshwater transitions are especially difficult."[51]
"Difficulty in detecting transitions between marine and freshwater systems may contribute to the paradigm that transitions occur infrequently. Detecting a transition requires sufficiently abundant extant descendants. Most immigrant cells are expected to go extinct locally due to ecological drift, just as most mutations are lost from a population due to genetic drift.[55] The probability of an immigrant avoiding extinction due to ecological drift (random births and deaths in a population [56]) like a mutation avoiding genetic drift, depends on the degree of selective advantage. For example, in populations of Escherichiacoli (∼3 × 107 cells,[57] a mutation conferring a 10% advantage appears an average of five times before it is established compared to a mutation with a 0.1% advantage which would need to appear 500 times to avoid extinction by drift.[58] In addition to overcoming ecological drift, the degree of selective advantage for cells migrating between marine and freshwater habitats would need to be strong enough to overcome any salinity-based disadvantages. Microorganisms that become established must also achieve sufficiently high population abundances to be reliably detected by current sequencing methods. As amplicon sequencing data sets accumulate from an increased diversity of environments and library size increases, the ability to detect transitions improves."[51]
Salinity preference
[edit]"Understanding if and how the evolutionary history of a species relates to its ecology is a fundamental question for biologists. For macroorganisms, phylogeny is often an ecologically meaningful way to classify organisms, as closely related taxa frequently have similar ecological characteristics (e.g., Silvertown et al., 2006; Donoghue, 2008) and functional traits (e.g., Cavender-Bares et al., 2009). In contrast, the phylogeny of microorganisms is generally considered to be an unreliable indicator of ecology because microbes can evolve quickly (Vasi et al., 1994) and engage in horizontal gene transfer (Snel et al., 2002). Nonetheless, most research into the ecology of microorganisms has relied on phylogenetic classification, and there is an accumulating body of evidence that shows phylogenetically-clustered taxa exhibit a substantial degree of ecological similarity, even at high levels of taxonomic organization (Philippot et al., 2010; Langille et al., 2013). For instance, ecological coherence (when members of a phylogenetic group share strategies or traits that distinguish them from other clades (Philippot et al., 2010)) has been demonstrated for the orders of α-proteobacteria, where members of each order are similar in multiple regards including habitat preference and genome size (Ettema and Andersson, 2009). However, there has been virtually no research into whether the responses of bacterial phylotypes to key environmental variables are phylogenetically conserved. An increased understanding of these relationships could lead to a more predictive understanding of how environmental variables shape bacterial community composition and could help explain global patterns in bacterial biodiversity.[59]
"Salinity has been proposed to be a major driver of phylogenetic bacterial community composition across the planet (Lozupone and Knight, 2007), and numerous studies in aquatic systems have found the relative abundance of high-level taxonomic groups (e.g., phylum, class) to correlate with salinity (e.g., del Giorgio and Bouvier, 2002; Herlemann et al., 2011). Though it has not yet been explored, one explanation for these patterns is that they reflect ecological coherence of salinity preferences. An increased understanding of the mechanisms by which salinity influences microbial communities is especially important given the widespread ongoing salinization of coastal habitats due to climate change and anthropogenic modification of the hydrologic cycle(Herbert et al., 2015). Tidal freshwater wetlands are particularly vulnerable, and sea-level rise is expected to cause saltwater intrusion into these ecosystems (Neubauer and Craft, 2009). To date, the biogeochemical responses of these systems to elevated salinity have been incongruous. For example, thermodynamic considerations have led many scientists to predict that methanogenesis, the dominant anaerobic pathway in organic matter degradation in tidal freshwater wetland soils, will be suppressed as salinity increases. This expectation is based on the fact that the elevated sulfate concentrations associated with more saline water allow sulfate-reducing bacteria (SRB) to outcompete methanogens. There are several reports in the literature that are consistent with this expectation (e.g., Weston et al., 2006; Chambers et al., 2011; Neubauer, 2013) but also some that are not (Weston et al., 2011; Hopfensperger et al., 2014). Along these same lines, increases in salinity have been reported to both increase (Weston et al., 2006; Craft, 2007) and decrease (Roache et al., 2006; Neubauer et al., 2013) decomposition rates in wetlands. These discordant responses may be driven by differences in the composition of the underlying microbial communities and their dynamic responses to altered salinity, which may be phylogenetically conserved. Thus, characterizing the salinity preferences of phylogenetic groups will enhance our ability to predict how a change in salinity will affect a given microbial community and its associated functions."[59]
References
[edit]- ^ a b Lepère, Cécile; Domaizon, Isabelle; Humbert, Jean-Francois; Jardillier, Ludwig; Hugoni, Mylène; Debroas, Didier (2019). "Diversity, spatial distribution and activity of fungi in freshwater ecosystems". PeerJ. 7: e6247. doi:10.7717/peerj.6247. PMC 6387782. PMID 30809429.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d Ren, Ze; Gao, Hongkai (2019). "Ecological networks reveal contrasting patterns of bacterial and fungal communities in glacier-fed streams in Central Asia". PeerJ. 7: e7715. doi:10.7717/peerj.7715. PMC 6753927. PMID 31576247.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e Battin, Tom J.; Besemer, Katharina; Bengtsson, Mia M.; Romani, Anna M.; Packmann, Aaron I. (2016). "The ecology and biogeochemistry of stream biofilms" (PDF). Nature Reviews Microbiology. 14 (4): 251–263. doi:10.1038/nrmicro.2016.15. PMID 26972916. S2CID 6573778.
- ^ De Carvalho, Carla C. C. R. (2018). "Marine Biofilms: A Successful Microbial Strategy with Economic Implications". Frontiers in Marine Science. 5. doi:10.3389/fmars.2018.00126. S2CID 4734366.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c d Wurzbacher, Christian; Fuchs, Andrea; Attermeyer, Katrin; Frindte, Katharina; Grossart, Hans-Peter; Hupfer, Michael; Casper, Peter; Monaghan, Michael T. (2017). "Shifts among Eukaryota, Bacteria, and Archaea define the vertical organization of a lake sediment". Microbiome. 5 (1): 41. doi:10.1186/s40168-017-0255-9. PMC 5385010. PMID 28388930.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Tranvik, Lars J.; Downing, John A.; Cotner, James B.; Loiselle, Steven A.; Striegl, Robert G.; Ballatore, Thomas J.; Dillon, Peter; Finlay, Kerri; Fortino, Kenneth; Knoll, Lesley B.; Kortelainen, Pirkko L.; Kutser, Tiit; Larsen, Soren.; Laurion, Isabelle; Leech, Dina M.; McCallister, S. Leigh; McKnight, Diane M.; Melack, John M.; Overholt, Erin; Porter, Jason A.; Prairie, Yves; Renwick, William H.; Roland, Fabio; Sherman, Bradford S.; Schindler, David W.; Sobek, Sebastian; Tremblay, Alain; Vanni, Michael J.; Verschoor, Antonie M.; et al. (2009). "Lakes and reservoirs as regulators of carbon cycling and climate". Limnology and Oceanography. 54 (6part2): 2298–2314. Bibcode:2009LimOc..54.2298T. doi:10.4319/lo.2009.54.6_part_2.2298. S2CID 54880187.
- ^ . doi:10.1130/0091-7613(1998)026 (inactive 2022-06-04).
{{cite journal}}: Cite journal requires|journal=(help); Missing or empty|title=(help)CS1 maint: DOI inactive as of June 2022 (link) - ^ Bastviken, D.; Tranvik, L. J.; Downing, J. A.; Crill, P. M.; Enrich-Prast, A. (2011). "Freshwater Methane Emissions Offset the Continental Carbon Sink". Science. 331 (6013): 50. Bibcode:2011Sci...331...50B. doi:10.1126/science.1196808. PMID 21212349. S2CID 31947846.
- ^ Fischer, Helmut; Sachse, Anke; Steinberg, Christian E. W.; Pusch, Martin (2002). "Differential retention and utilization of dissolved organic carbon by bacteria in river sediments". Limnology and Oceanography. 47 (6): 1702–1711. Bibcode:2002LimOc..47.1702F. doi:10.4319/lo.2002.47.6.1702. S2CID 97827704.
- ^ Haglund, Ann-Louise; Lantz, Peter; Tã¶Rnblom, Erik; Tranvik, Lars (2003). "Depth distribution of active bacteria and bacterial activity in lake sediment". FEMS Microbiology Ecology. 46 (1): 31–38. doi:10.1016/S0168-6496(03)00190-9. PMID 19719580.
- ^ Nealson, Kenneth H. (1997). "SEDIMENT BACTERIA: Who's There, What Are They Doing, and What's New?". Annual Review of Earth and Planetary Sciences. 25: 403–434. Bibcode:1997AREPS..25..403N. doi:10.1146/annurev.earth.25.1.403. PMID 11540735.
- ^ Megonigal, J. Patrick; Neubauer, Scott C. (2019). "Biogeochemistry of Tidal Freshwater Wetlands". Coastal Wetlands. pp. 641–683. doi:10.1016/B978-0-444-63893-9.00019-8. ISBN 9780444638939. S2CID 131221695.
- ^ Maerki, Martin; Müller, Beat; Dinkel, Christian; Wehrli, Bernhard (2009). "Mineralization pathways in lake sediments with different oxygen and organic carbon supply". Limnology and Oceanography. 54 (2): 428–438. Bibcode:2009LimOc..54..428M. doi:10.4319/lo.2009.54.2.0428. S2CID 130983546.
- ^ Melton, Emily D.; Swanner, Elizabeth D.; Behrens, Sebastian; Schmidt, Caroline; Kappler, Andreas (2014). "The interplay of microbially mediated and abiotic reactions in the biogeochemical Fe cycle". Nature Reviews Microbiology. 12 (12): 797–808. doi:10.1038/nrmicro3347. PMID 25329406. S2CID 24058676.
- ^ Frindte, Katharina; Allgaier, Martin; Grossart, Hans-Peter; Eckert, Werner (2015). "Microbial Response to Experimentally Controlled Redox Transitions at the Sediment Water Interface". PLOS ONE. 10 (11): e0143428. Bibcode:2015PLoSO..1043428F. doi:10.1371/journal.pone.0143428. PMC 4657962. PMID 26599000. S2CID 15406422.
- ^ a b Lloyd, Karen G.; Schreiber, Lars; Petersen, Dorthe G.; Kjeldsen, Kasper U.; Lever, Mark A.; Steen, Andrew D.; Stepanauskas, Ramunas; Richter, Michael; Kleindienst, Sara; Lenk, Sabine; Schramm, Andreas; Jørgensen, Bo Barker (2013). "Predominant archaea in marine sediments degrade detrital proteins". Nature. 496 (7444): 215–218. Bibcode:2013Natur.496..215L. doi:10.1038/nature12033. PMID 23535597. S2CID 4395910.
- ^ Miskin, I.; Rhodes, G.; Lawlor, K.; Saunders, J. R.; Pickup, R. W. (1998). "Bacteria in post-glacial freshwater sediments". Microbiology. 144 (9): 2427–2439. doi:10.1099/00221287-144-9-2427. PMID 9782490.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Li, Shengkang; Xiao, Xiang; Yin, Xuebin; Wang, Fengping (2006). "Bacterial community along a historic lake sediment core of Ardley Island, west Antarctica". Extremophiles. 10 (5): 461–467. doi:10.1007/s00792-006-0523-2. PMID 16715185. S2CID 22447365.
- ^ a b Ye, Wenjin; Liu, Xianglong; Lin, Shengqin; Tan, Jing; Pan, Jianliang; Li, Daotang; Yang, Hong (2009). "The vertical distribution of bacterial and archaeal communities in the water and sediment of Lake Taihu". FEMS Microbiology Ecology. 70 (2): 263–276. doi:10.1111/j.1574-6941.2009.00761.x. PMID 19744240.
- ^ Vuillemin, Aurèle; Ariztegui, Daniel; PASADO Science Team (2013). "Geomicrobiological investigations in subsaline maar lake sediments over the last 1500 years". Quaternary Science Reviews. 71: 119–130. Bibcode:2013QSRv...71..119V. doi:10.1016/j.quascirev.2012.04.011.
- ^ Xiong, Wei; Xie, Ping; Wang, Shengrui; Niu, Yuan; Yang, Xi; Chen, Wenjie (2014). "Sources of organic matter affect depth-related microbial community composition in sediments of Lake Erhai, Southwest China". Journal of Limnology. 73. doi:10.4081/jlimnol.2014.1106.
- ^ Cite error: The named reference
Nealson1997was invoked but never defined (see the help page). - ^ Spring, S. (2000). "Identification and characterization of ecologically significant prokaryotes in the sediment of freshwater lakes: Molecular and cultivation studies". FEMS Microbiology Reviews. 24 (5): 573–590. doi:10.1016/S0168-6445(00)00046-2. PMID 11077151.
- ^ Briée, Céline; Moreira, David; López-García, Purificación (2007). "Archaeal and bacterial community composition of sediment and plankton from a suboxic freshwater pond". Research in Microbiology. 158 (3): 213–227. doi:10.1016/j.resmic.2006.12.012. PMID 17346937.
- ^ Lipp, Julius S.; Morono, Yuki; Inagaki, Fumio; Hinrichs, Kai-Uwe (2008). "Significant contribution of Archaea to extant biomass in marine subsurface sediments". Nature. 454 (7207): 991–994. Bibcode:2008Natur.454..991L. doi:10.1038/nature07174. PMID 18641632. S2CID 4316347.
- ^ a b Lloyd, Karen G.; May, Megan K.; Kevorkian, Richard T.; Steen, Andrew D. (2013). "Meta-Analysis of Quantification Methods Shows that Archaea and Bacteria Have Similar Abundances in the Subseafloor". Applied and Environmental Microbiology. 79 (24): 7790–7799. doi:10.1128/AEM.02090-13. PMC 3837824. PMID 24096423. S2CID 12431461.
- ^ Borrel, Guillaume; Lehours, Anne-Catherine; Crouzet, Olivier; Jézéquel, Didier; Rockne, Karl; Kulczak, Amélie; Duffaud, Emilie; Joblin, Keith; Fonty, Gérard (2012). "Stratification of Archaea in the Deep Sediments of a Freshwater Meromictic Lake: Vertical Shift from Methanogenic to Uncultured Archaeal Lineages". PLOS ONE. 7 (8): e43346. Bibcode:2012PLoSO...743346B. doi:10.1371/journal.pone.0043346. PMC 3424224. PMID 22927959. S2CID 10812384.
- ^ Zhang, Jingxu; Yang, Yuyin; Zhao, Lei; Li, Yuzhao; Xie, Shuguang; Liu, Yong (2015). "Distribution of sediment bacterial and archaeal communities in plateau freshwater lakes". Applied Microbiology and Biotechnology. 99 (7): 3291–3302. doi:10.1007/s00253-014-6262-x. PMID 25432677. S2CID 6772606.
- ^ Rodrigues, Thiago; Catão, Elisa; Bustamante, Mercedes M. C.; Quirino, Betania F.; Kruger, Ricardo H.; Kyaw, Cynthia M. (2014). "Seasonal Effects in a Lake Sediment Archaeal Community of the Brazilian Savanna". Archaea. 2014: 1–9. doi:10.1155/2014/957145. PMC 4131120. PMID 25147480. S2CID 15084103.
- ^ Gächter, René; Meyer, Joseph S.; Mares, Antonin (1988). "Contribution of bacteria to release and fixation of phosphorus in lake sediments". Limnology and Oceanography. 33 (6part2): 1542–1558. Bibcode:1988LimOc..33.1542G. doi:10.4319/lo.1988.33.6part2.1542. S2CID 86461426.
- ^ Chan, On Chim; Claus, Peter; Casper, Peter; Ulrich, Andreas; Lueders, Tillmann; Conrad, Ralf (2005). "Vertical distribution of structure and function of the methanogenic archaeal community in Lake Dagow sediment". Environmental Microbiology. 7 (8): 1139–1149. doi:10.1111/j.1462-2920.2005.00790.x. PMID 16011751.
- ^ Cite error: The named reference
Haglund2003was invoked but never defined (see the help page). - ^ Polymenakou, Paraskevi N.; Fragkioudaki, Glykeria; Tselepides, Anastasios (2007). "Bacterial and organic matter distribution in the sediments of the Thracian Sea (NE Aegean Sea)". Continental Shelf Research. 27 (17): 2187–2197. Bibcode:2007CSR....27.2187P. doi:10.1016/j.csr.2007.05.003.
- ^ Lomstein, Bente Aa.; Langerhuus, Alice T.; d'Hondt, Steven; Jørgensen, Bo B.; Spivack, Arthur J. (2012). "Endospore abundance, microbial growth and necromass turnover in deep sub-seafloor sediment". Nature. 484 (7392): 101–104. Bibcode:2012Natur.484..101L. doi:10.1038/nature10905. PMID 22425999. S2CID 205228035.
- ^ Xie, S.; Lipp, J. S.; Wegener, G.; Ferdelman, T. G.; Hinrichs, K.-U. (2013). "Turnover of microbial lipids in the deep biosphere and growth of benthic archaeal populations". Proceedings of the National Academy of Sciences. 110 (15): 6010–6014. Bibcode:2013PNAS..110.6010X. doi:10.1073/pnas.1218569110. PMC 3625344. PMID 23530229. S2CID 22102448.
- ^ Hoehler, Tori M.; Jørgensen, Bo Barker (2013). "Microbial life under extreme energy limitation". Nature Reviews Microbiology. 11 (2): 83–94. doi:10.1038/nrmicro2939. hdl:2060/20150018056. PMID 23321532. S2CID 461070.
- ^ Tanentzap, Andrew J (19 November 2019) Freshwater lakes already emit a quarter of global carbon – and climate change could double that The Conversation.
- ^ Grossart, Hans-Peter (2010). "Ecological consequences of bacterioplankton lifestyles: Changes in concepts are needed". Environmental Microbiology Reports. 2 (6): 706–714. doi:10.1111/j.1758-2229.2010.00179.x. PMID 23766274.
- ^ Cole, J. J. (1982). "Interactions Between Bacteria and Algae in Aquatic Ecosystems". Annual Review of Ecology and Systematics. 13: 291–314. doi:10.1146/annurev.es.13.110182.001451.
- ^ Koblížek, Michal (2015). "Ecology of aerobic anoxygenic phototrophs in aquatic environments". FEMS Microbiology Reviews. 39 (6): 854–870. doi:10.1093/femsre/fuv032. PMID 26139241.
- ^ Mizuno, Carolina Megumi; Rodriguez-Valera, Francisco; Ghai, Rohit (2015). "Genomes of Planktonic Acidimicrobiales: Widening Horizons for Marine Actinobacteria by Metagenomics". mBio. 6 (1). doi:10.1128/mBio.02083-14. PMC 4337565. PMID 25670777.
- ^ Salcher, Michaela M.; Neuenschwander, Stefan M.; Posch, Thomas; Pernthaler, Jakob (2015). "The ecology of pelagic freshwater methylotrophs assessed by a high-resolution monitoring and isolation campaign". The ISME Journal. 9 (11): 2442–2453. doi:10.1038/ismej.2015.55. PMC 4611508. PMID 25942006.
- ^ Martiny, J. B. H.; Jones, S. E.; Lennon, J. T.; Martiny, A. C. (2015). "Microbiomes in light of traits: A phylogenetic perspective". Science. 350 (6261): aac9323. Bibcode:2015Sci...350.9323M. doi:10.1126/science.aac9323. PMID 26542581. S2CID 1460155.
- ^ a b Dupont, Chris L.; Larsson, John; Yooseph, Shibu; Ininbergs, Karolina; Goll, Johannes; Asplund-Samuelsson, Johannes; McCrow, John P.; Celepli, Narin; Allen, Lisa Zeigler; Ekman, Martin; Lucas, Andrew J.; Hagström, Åke; Thiagarajan, Mathangi; Brindefalk, Björn; Richter, Alexander R.; Andersson, Anders F.; Tenney, Aaron; Lundin, Daniel; Tovchigrechko, Andrey; Nylander, Johan A. A.; Brami, Daniel; Badger, Jonathan H.; Allen, Andrew E.; Rusch, Douglas B.; Hoffman, Jeff; Norrby, Erling; Friedman, Robert; Pinhassi, Jarone; Venter, J. Craig; Bergman, Birgitta (2014). "Functional Tradeoffs Underpin Salinity-Driven Divergence in Microbial Community Composition". PLOS ONE. 9 (2): e89549. Bibcode:2014PLoSO...989549D. doi:10.1371/journal.pone.0089549. PMC 3937345. PMID 24586863.
- ^ Cite error: The named reference
Global patterns in bacterial diverswas invoked but never defined (see the help page). - ^ Thompson, Luke R.; Sanders, Jon G.; McDonald, Daniel; Amir, Amnon; Ladau, Joshua; Locey, Kenneth J.; Prill, Robert J.; Tripathi, Anupriya; Gibbons, Sean M.; Ackermann, Gail; Navas-Molina, Jose A.; Janssen, Stefan; Kopylova, Evguenia; Vázquez-Baeza, Yoshiki; González, Antonio; Morton, James T.; Mirarab, Siavash; Zech Xu, Zhenjiang; Jiang, Lingjing; Haroon, Mohamed F.; Kanbar, Jad; Zhu, Qiyun; Jin Song, Se; Kosciolek, Tomasz; Bokulich, Nicholas A.; Lefler, Joshua; Brislawn, Colin J.; Humphrey, Gregory; Owens, Sarah M.; et al. (2017). "A communal catalogue reveals Earth's multiscale microbial diversity". Nature. 551 (7681): 457–463. Bibcode:2017Natur.551..457T. doi:10.1038/nature24621. PMC 6192678. PMID 29088705.
- ^ Herlemann, Daniel PR; Labrenz, Matthias; Jürgens, Klaus; Bertilsson, Stefan; Waniek, Joanna J.; Andersson, Anders F. (2011). "Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea". The ISME Journal. 5 (10): 1571–1579. doi:10.1038/ismej.2011.41. PMC 3176514. PMID 21472016.
- ^ Fortunato, Caroline S.; Crump, Byron C. (2015). "Microbial Gene Abundance and Expression Patterns across a River to Ocean Salinity Gradient". PLOS ONE. 10 (11): e0140578. Bibcode:2015PLoSO..1040578F. doi:10.1371/journal.pone.0140578. PMC 4633275. PMID 26536246.
- ^ Logares, Ramiro; Lindström, Eva S.; Langenheder, Silke; Logue, Jürg B.; Paterson, Harriet; Laybourn-Parry, Johanna; Rengefors, Karin; Tranvik, Lars; Bertilsson, Stefan (2013). "Biogeography of bacterial communities exposed to progressive long-term environmental change". The ISME Journal. 7 (5): 937–948. doi:10.1038/ismej.2012.168. PMC 3635229. PMID 23254515.
- ^ Logares, Ramiro; Bråte, Jon; Bertilsson, Stefan; Clasen, Jessica L.; Shalchian-Tabrizi, Kamran; Rengefors, Karin (2009). "Infrequent marine–freshwater transitions in the microbial world". Trends in Microbiology. 17 (9): 414–422. doi:10.1016/j.tim.2009.05.010. PMID 19726194.
- ^ a b c d e f g h Paver, Sara F.; Muratore, Daniel; Newton, Ryan J.; Coleman, Maureen L. (2018). "Reevaluating the Salty Divide: Phylogenetic Specificity of Transitions between Marine and Freshwater Systems". mSystems. 3 (6). doi:10.1128/mSystems.00232-18. PMC 6234284. PMID 30443603.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Logares, R.; Brate, J.; Heinrich, F.; Shalchian-Tabrizi, K.; Bertilsson, S. (2010). "Infrequent Transitions between Saline and Fresh Waters in One of the Most Abundant Microbial Lineages (SAR11)". Molecular Biology and Evolution. 27 (2): 347–357. doi:10.1093/molbev/msp239. PMID 19808864.
- ^ Zwart, G.; Crump, BC; Kamst-Van Agterveld, MP; Hagen, F.; Han, SK (2002). "Typical freshwater bacteria: An analysis of available 16S rRNA gene sequences from plankton of lakes and rivers". Aquatic Microbial Ecology. 28: 141–155. doi:10.3354/ame028141.
- ^ a b Newton, R. J.; Jones, S. E.; Eiler, A.; McMahon, K. D.; Bertilsson, S. (2011). "A Guide to the Natural History of Freshwater Lake Bacteria". Microbiology and Molecular Biology Reviews. 75 (1): 14–49. doi:10.1128/MMBR.00028-10. PMC 3063352. PMID 21372319.
- ^ Hubbell, Stephen P. (29 April 2001). The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32). ISBN 0691021287.
- ^ Fodelianakis, Stilianos; Valenzuela-Cuevas, Adriana; Barozzi, Alan; Daffonchio, Daniele (2020). "Direct quantification of ecological drift at the population level in synthetic bacterial communities". The ISME Journal. 15 (1): 55–66. doi:10.1038/s41396-020-00754-4. PMC 7852547. PMID 32855435.
- ^ Lenski, Richard E.; Rose, Michael R.; Simpson, Suzanne C.; Tadler, Scott C. (1991). "Long-Term Experimental Evolution in Escherichia coli. I. Adaptation and Divergence During 2,000 Generations". The American Naturalist. 138 (6): 1315–1341. doi:10.1086/285289. S2CID 83996233.
- ^ Elena, Santiago F.; Lenski, Richard E. (2003). "Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation". Nature Reviews Genetics. 4 (6): 457–469. doi:10.1038/nrg1088. PMID 12776215. S2CID 209727.
- ^ a b Morrissey, Ember M.; Franklin, Rima B. (2015). "Evolutionary history influences the salinity preference of bacterial taxa in wetland soils". Frontiers in Microbiology. 6: 1013. doi:10.3389/fmicb.2015.01013. PMC 4591843. PMID 26483764.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Freshwater plankton
[edit]

- Freshwater phytoplankton
- Lake plankton
Freshwater plankton are plankton defined by their habit as occurring within freshwater ecosystems. They can be divided further, into sub habitats, as:[1]
- limnoplankton: lake plankton
- heleoplankton: pond plankton
- potamoplankton: river plankton, plankton of running water, sometimes called rheoplankton
Overview
[edit]Freshwater plankton is defined by its habitat or environment as plankton that lives in freshwater. It can be contrasted with marine plankton, which is plankton that lives in marine environments, such as the saltwaters of oceans and the brackish waters of estuaries.
"Freshwater ecosystems harbor a vast diversity of micro-eukaryotes (rotifers, crustaceans and protists), and such diverse taxonomic groups play important roles in ecosystem functioning and services. Unfortunately, freshwater ecosystems and biodiversity therein are threatened by many environmental stressors, particularly those derived from intensive human activities such as chemical pollution. In the past several decades, significant efforts have been devoted to halting biodiversity loss to recover services and functioning of freshwater ecosystems. Biodiversity monitoring is the first and a crucial step towards diagnosing pollution impacts on ecosystems and making conservation plans. Yet, bio-monitoring of ubiquitous micro-eukaryotes is extremely challenging, owing to many technical issues associated with micro-zooplankton such as microscopic size, fuzzy morphological features, and extremely high biodiversity."[2]
"Among various types of ecosystems, freshwater ecosystems provide unique habitats, supporting a high level of biodiversity. Freshwater ecosystems occupy only approximately 0.8% of the Earth’s surface but support almost 6% of all known species.[3][2]

Phytoplankon
[edit]
"Freshwater phytoplankton is well represented by species of most of the major divisions of algae that have a planktonic component. Therefore, a plankton sample from a lake may contain cyanobacteria, Chlorophyta, Chrysophyta, and dinoflagellates. Euglenophyta, which can usually be found in small ponds, are not common members of the freshwater phytoplankton (Round 1981). A large number of species of Chlorophyta and Cyanophyta are found in freshwater, but not in seawater. Coccolithophores, which are characteristic of marine plankton, are rarely observed in freshwaters."[4]

- freshwater phytoplankton
-
Oscillatoria princeps, a filamentous cyanobacterium found in ponds
-
Pediastrum duplex, a fresh water green algae
-
Volvox, a genus of chlorophyte green algae which form spherical colonies of up to 50,000 cells
-
Freshwater diatom from a pond in Moscow
-
Prototheca wickerhamii, an infectious alga
-
Euglenophyta – usually found in small ponds
-
Dinobryon divergens (golden algae)
- zooplankton
Common types of lake zooplankton are rotifers, water fleas and copepods.[8]
Copepods are typically 1 to 2 mm long with a teardrop-shaped bodies. Like all crustaceans, their bodies are divided into three sections: head, thorax, and abdomen, with two pairs of antennae; the first pair is often long and prominent. They have a tough exoskeleton made of calcium carbonate and usually have a single red eye in the centre of their transparent head.[9] About 2,800 species are known to live in freshwater,[10] and they are usually the dominant members of the zooplankton.[11] Freshwater copepods of the Cyclops genus are the intermediate host of Dracunculus medinensis, the Guinea worm nematode that causes dracunculiasis disease in humans. This disease may be close to being eradicated through efforts at the U.S. Centers for Disease Control and Prevention and the World Health Organization.[12]
| External videos | |
|---|---|
- freshwater bacterioplankton
- saline lakes
Saline lakes are often shallow and well mixed. They respond quickly to even small climatic changes, and can sensitively record climatic and geological changes.[13]

- Freshwater habitats of plankton
Terrestrial aquatic habitats include lakes and reservoirs, rivers and streams, farm dams, ponds and swamps. Human formed reservoirs are more numerous than natural lakes. Evaporation dries ponds, rainfall floods fields, some streams flow only after rain.[14]
Inland waters are generally fresh, with low concentrations of dissolved salts compared to marine and estuarine environments. Some inland waters, like salt lakes, can be saltier than seawater. But there is no clear demarcation between freshwater and saltwater habits, so demarcations that are set for convenience will be somewhat arbitrary.[13] Williams (1980) has acknowledged this issue at some length, and then, as a practical matter, defined freshwater as having a salinity "less than 2 g/L of dissolved salts".[13] The composition of salts also varies. Salts from lowland areas with low rainfall and high evaporation are often like seawater salts, dominated by sodium and chloride. Salts from upland headwater streams and reservoirs are often dominated by calcium and magnesium bicarbonates.[14]
- Rivers
Rivers can overflow during flood events across floodplains, depositing large quantities of sediment and nutrients on the plains. Stream flows can be sustained by groundwater drainage during droughts. Upland rivers and streams are typically shallow, dropping steeply with high velocity flows keeping waters well mixed. When rivers reach lowlands the gradients ease, and rivers can become broad and meandering. Or they might split into anabranches and distributary channels and terminate in wetlands. Lowland rivers can enter natural ponds and constructed weirs and slow down, becoming deeper and more lake-like.[15] [14]
NEEDS PARAPHRASING...
"Flowing river ecosystems are generally not good habitats for plankton, because the organisms entrained within the water column are continually displaced downstream. However, some of the larger lowland rivers may develop their own riverine phytoplankton communities that develop within parcels of water as these traverse the length of the river. Most algal growth in smaller, shallower, faster flowing streams, however, is confined to clumps of filamentous algae attached to a secure substrate to prevent themselves from being washed away, and to films of microscopic algae coating the surfaces of rocks, mud, sticks and aquatic macrophytes. These algae obtain the substances they require to sustain their growth as the water flows over them. The weir pools and ponded sections of lowland rivers and streams may, however, become suitable habits for phytoplankton to form blooms. Some rivers also have small embayments, inlets or backwater areas where water movement may be mal. These areas - known as head runes' -are also areas where phytoplanlitort can develop (Mitrovic It h. 20011."[14]
"Lakes, reservoir, farm dams, ponds, billabongs and wetlands are characterised by prolonged resi-dence times of the water they contain and the lim-ited mixing of water within them-apart from that caused by wind-driven currents and internal-heat-transfer processes. Damper lakes and reset ovirs undergo strong thermal stratification during the warmer months of the year, caused by the preferen-til solar heating of the surface waters. Water den-sity decreases as temperature increases, so warm water overlies colder water and creates horizontal density gradients that resist vertical mixing and enhance the stability of the water column. Chemi-cal and biological demand for oxygen in deeper regions, accompanied by limited replenishment from the surface due to the lack of vertical mixing, can lead to very low oxygen levels in deep lake waters (Smith el al. 201I). Deoxygenation of the deeper waters has major effects on the chemistry of other substances, especially nutrients, which can be mobilised from the lake sediments under such conditions The thermal stratification and mixing regimes of lakes and reservoirs influences water column stability, nutrient and light availability at different times of the year and, coreequently, the plankton community."[14]
"Farm dams are often very turbid environments, so lack of light within the water column may limit phytoplankton growth. These, aSd other small ponds, are often typified by high amounts of organic substances in the water, which is often thought M favour certain kinds of motile unicellu-lar algae known as euglenoids (Section 5.6). Wet-lands and billabongs are shallow and much of the submerged area is occupied by aquatic macro-phyla and large macroalgae, known as charo-phyla, tha grow from the sediments. These macropbyten and algae that grow attached to them (termed epiphytes) may compete with phytoplank-ton for light and nutrients, so that wetlands may not be good habitats for phytoplankton and zoo-plankton. Shallow water bodies may be clear water, xnacrophyte-dominated systems, or MM., nut-ti-ent-enriched, phytoplankton-dominated systems (Scheffer 1998) In very dry regions, suitable Mon dams are often very turbid environments, m lack of light within the water column may limit phytoplankton growth. These, aSd other small ponds, are often typified by high amounts of organic substances in the water, which is often thought M favour certain kinds of motile unicellu-lar algae known as euglenoids (Section 5.6). Wet-lands and billabongs are shallow and much of the submerged area is occupied by aquatic macro-phyla and large macroalgae, known as charo-phyla, tha grow from the sediments. These macropbyten and algae that grow attached to them (termed epiphytes) may compete with phytoplank-ton for light and nutrients, so that wetlands may not be good habitats for phytoplankton and zoo-plankton. Shallow water bodies may be clear water, xnacrophyte-dominated systems, or MM., nut-ti-ent-enriched, phytoplankton-dominated systems (Scheffer 1998) In very dry regions, suitable habitats for plankton may only he present for short periods following brief periods of rairdall, and ponds may dry up rapidly often for long periods. The plankton associated with these environment have evolved strategies that still enable them to thrive in such harsh environments."[14]
- ponds
Plankton blooms
[edit]

Human impact
[edit]

"Freshwater ecosystems provide irreplaceable goods and services for human beings, such as drinking and irrigation water, food, creation, and regulation of micro-climate [4,5]."[2]
"However, many factors, particularly those derived from anthropogenic activities such as water pollution and invasive species, have largely degraded freshwater ecosystems over the past several decades [2,6]. Freshwater ecosystems such as rivers and inland lakes are among the most threatened ecosystems on the Earth [4,7]. As a result, biodiversity loss in freshwater ecosystems is much faster than that in terrestrial counterparts [8]. Even worse, biodiversity loss in threaten freshwater ecosystems have not slowed down in recent years [9], despite the fact that great effort has been placed on maintaining or recovering biodiversity in freshwater ecosystems. These efforts have been largely unsuccessful due to frequent disturbance derived from increasing anthropogenic activities and knowledge gaps on biodiversity in freshwater ecosystems [10]."[2]
"Indeed, biodiversity loss in freshwater ecosystem is likely much more severe than we have realized, as biological response to disturbance in freshwater ecosystems is not completely known, especially on the widespread but hidden microscopic taxa such as zooplankton [3]. A large body of scientific literature has illustrated the biodiversity loss of macro-eukaryotes under human activity disturbance, such as fishes, amphibians, mollusks and crustaceans 11, [12], [13], 14. Nevertheless, studies on biodiversity loss dynamics of micro-eukaryotes are rare [15]. In terms of monitoring and conservation priority, better-known macro-eukaryotes have drawn more attention than micro-eukaryotes such as microscopic zooplankton [16]. Indeed, the highly neglected microscopic zooplankton play vital ecological roles in aquatic food-webs, such as linking phytoplankton and bacteria to high trophic levels such as fish [17]. The protection and recovery of overlooked microscopic zooplankton biodiversity largely determine the conservation of biodiversity at high trophic levels, as well as the integration and functioning of freshwater ecosystems."[2]
NEEDS PARAPHRASING...
"Freshwater biodiversity is the over-riding conservation priority during the International Decade for Action – ‘Water for Life’ – 2005 to 2015. Fresh water makes up only 0.01% of the World's water and approximately 0.8% of the Earth's surface, yet this tiny fraction of global water supports at least 100000 species out of approximately 1.8 million – almost 6% of all described species. Inland waters and freshwater biodiversity constitute a valuable natural resource, in economic, cultural, aesthetic, scientific and educational terms. Their conservation and management are critical to the interests of all humans, nations and governments. Yet this precious heritage is in crisis. Fresh waters are experiencing declines in biodiversity far greater than those in the most affected terrestrial ecosystems, and if trends in human demands for water remain unaltered and species losses continue at current rates, the opportunity to conserve much of the remaining biodiversity in fresh water will vanish before the ‘Water for Life’ decade ends in 2015. Why is this so, and what is being done about it? This article explores the special features of freshwater habitats and the biodiversity they support that makes them especially vulnerable to human activities. We document threats to global freshwater biodiversity under five headings: overexploitation; water pollution; flow modification; destruction or degradation of habitat; and invasion by exotic species. Their combined and interacting influences have resulted in population declines and range reduction of freshwater biodiversity worldwide. Conservation of biodiversity is complicated by the landscape position of rivers and wetlands as ‘receivers’ of land-use effluents, and the problems posed by endemism and thus non-substitutability. In addition, in many parts of the world, fresh water is subject to severe competition among multiple human stakeholders. Protection of freshwater biodiversity is perhaps the ultimate conservation challenge because it is influenced by the upstream drainage network, the surrounding land, the riparian zone, and – in the case of migrating aquatic fauna – downstream reaches. Such prerequisites are hardly ever met."[3]
- Potential indicative roles of zooplankton in freshwater ecosystems
"Zooplankton include diverse taxa such as protists, rotifers, copepods and cladocerans, many of which are microscopic [18]. Multiple studies have made a consistent and crucial realization that zooplankton taxa are rapid responders to many environmental stressors, such as hydrological changes, climate changes and anthropogenic activity-induced water pollution [19,20]. Specifically, previous laboratory or field studies have indicated that zooplankton communities were significantly impacted by excessive loading of nutrients [15,19,21], and also negatively affected by microplastics [22], pesticides [23], and pharmaceuticals and personal care products (PPCPs) [24]. As such, researchers have identified their usefulness as ecological indicators to water pollution. For instances, rotifers are used to diagnose ecological impacts of freshwater toxicants, such as endocrine disruptors, bioconcentration of lead, and nanoparticles toxicity [25]. Yang et al. [26] indicated that zooplankton communities could be used to predict ecological thresholds of ammonia nitrogen. Payne et al. [27] listed and recommended seven key reasons for the use of protists as good bio-indicators in aquatic ecosystems. Azevêdo et al. [28] showed that zooplankton communities played a complementary role to macroinvertebrates in indicating variation of the trophic status of waters. Thus, bio-monitoring zooplankton communities has become a widely accepted and irreplaceable aspect in ecological conservation and management of aquatic ecosystems."[2]
References
[edit]- ^ S., Reynolds, Colin (2006). Ecology of phytoplankton. Cambridge: Cambridge University Press. p. 2. ISBN 978-0511191817. OCLC 76416312.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e f Xiong, Wei; Huang, Xuena; Chen, Yiyong; Fu, Ruiying; Du, Xun; Chen, Xingyu; Zhan, Aibin (2020). "Zooplankton biodiversity monitoring in polluted freshwater ecosystems: A technical review". Environmental Science and Ecotechnology. 1: 100008. doi:10.1016/j.ese.2019.100008. S2CID 214311725.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Kilham, Peter; Hecky, Robert E. (1988). "Comparative ecology of marine and freshwater phytoplankton1". Limnology and Oceanography. 33 (4part2): 776–795. Bibcode:1988LimOc..33..776K. doi:10.4319/lo.1988.33.4part2.0776. hdl:2027.42/109932.
- ^ Kilham, Peter; Hecky, Robert E. (1988). "Comparative ecology of marine and freshwater phytoplankton1". Limnology and Oceanography. 33 (4part2): 776–795. Bibcode:1988LimOc..33..776K. doi:10.4319/lo.1988.33.4part2.0776. hdl:2027.42/109932.
- ^ Hoshina, Ryo; Hayakawa, Masashi M.; Kobayashi, Mayumi; Higuchi, Rina; Suzaki, Toshinobu (2020). "Pediludiella daitoensis gen. Et sp. Nov. (Scenedesmaceae, Chlorophyceae), a large coccoid green alga isolated from a Loxodes ciliate". Scientific Reports. 10 (1): 628. Bibcode:2020NatSR..10..628H. doi:10.1038/s41598-020-57423-x. PMC 6971069. PMID 31959793.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Gontcharov AA, Marin BA, Melkonian MA (January 2003). "Molecular phylogeny of conjugating green algae (Zygnemophyceae, Streptophyta) inferred from SSU rDNA sequence comparisons". J. Mol. Evol. 56 (1): 89–104. doi:10.1007/s00239-002-2383-4. PMID 12569426. S2CID 35734083.
- ^ See the NCBI webpage on Cosmarium. Data extracted from the "NCBI taxonomy resources". National Center for Biotechnology Information. Retrieved 2007-03-19.
- ^ Zooplankton taxonomy: The three major groups of freshwater zooplankton Plankton Web. Accessed 26 November 2020. Updated 25 January 2013.
- ^ Robert D. Barnes (1982). Invertebrate Zoology. Philadelphia, Pennsylvania: Holt-Saunders International. pp. 683–692. ISBN 978-0-03-056747-6.
- ^ Geoff A. Boxhall; Danielle Defaye (2008). "Global diversity of copepods (Crustacea: Copepoda) in freshwater". Hydrobiologia. 595 (1): 195–207. doi:10.1007/s10750-007-9014-4. S2CID 31727589.
- ^ Johannes Dürbaum; Thorsten Künnemann (November 5, 1997). "Biology of Copepods: An Introduction". Carl von Ossietzky University of Oldenburg. Archived from the original on May 26, 2010. Retrieved December 8, 2009.
- ^ "This Species is Close to Extinction and That's a Good Thing". Time. January 23, 2015. Archived from the original on May 24, 2015. Retrieved May 31, 2015.
- ^ a b c Williams, W. D. (1981). "Inland salt lakes: An introduction". Salt Lakes. pp. 1–14. doi:10.1007/978-94-009-8665-7_1. ISBN 978-94-009-8667-1.
- ^ a b c d e f Suthers, I.M., Redden, A.M., Bowling, L., Kobayashi, T. and Rissik, D. (2009) "Plankton processes and the environment". In: Iain M Suthers, David Rissik (Eds.) Plankton: A Guide to Their Ecology and Monitoring for Water Quality, CSIRO Publishing, pages 21–36. ISBN 9780643099432.
- ^ Mitrovic, S. M.; Oliver, R. L.; Rees, C.; Bowling, L. C.; Buckney, R. T. (2003). "Critical flow velocities for the growth and dominance of Anabaena circinalis in some turbid freshwater rivers". Freshwater Biology. 48: 164–174. doi:10.1046/j.1365-2427.2003.00957.x.
Sources
[edit]- Suthers, Iain M.; Rissik, David (22 May 2009). Plankton: A Guide to Their Ecology and Monitoring for Water Quality. ISBN 9780643099432.
- Thorp, James H.; Covich, Alan P. (2010). Ecology and Classification of North American Freshwater Invertebrates. ISBN 9780123748553.
Piper
[edit]Phytoplankton defenses against zooplankton grazers
[edit]
against generalist and specialist zooplankton grazers

Predation is generally largest on small-sized phytoplankton. Sedimentation is usually highest for large cells, colonies, and aggregates that have no buoyancy control, and small cells that have rather high specific mass (diatoms). Competition is mostly strongest on larger cells and colonies that have a less favorable surface-to-volume ratio, and thus lower growth rates.[1]
← In the diagram on the left, small-celled, fast-growing cells suffer most from predation (1) but have in general low sinking loss, and because of favorable surface-to-volume ratio generally experiences less competition. Diatoms can only prevail in a mixed water column (4). Large, sometimes armored cells or colonies have lower growth rates, experience higher sinking rates, but are protected against grazing (2). Some of the slow-growing colonies or filaments possess the capacity of buoyancy control. The gray arrows indicate potential phenotypic plasticity.[1]
Carbon cycle overview
[edit]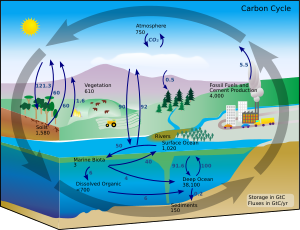

FROM: Carbon...
Under terrestrial conditions, conversion of one element to another is very rare. Therefore, the amount of carbon on Earth is effectively constant. Thus, processes that use carbon must obtain it from somewhere and dispose of it somewhere else. The paths of carbon in the environment form the carbon cycle. For example, photosynthetic plants draw carbon dioxide from the atmosphere (or seawater) and build it into biomass, as in the Calvin cycle, a process of carbon fixation. Some of this biomass is eaten by animals, while some carbon is exhaled by animals as carbon dioxide. The carbon cycle is considerably more complicated than this short loop; for example, some carbon dioxide is dissolved in the oceans; if bacteria do not consume it, dead plant or animal matter may become petroleum or coal, which releases carbon when burned.[2][3] Carbon occurs in all known organic life and is the basis of organic chemistry.
Fast and slow cycles
[edit]


Most carbon is sequestered "in the crustal rocks, about 100,000 times more than is in the atmosphere, but does not participate in the fast carbon cycle. "Most of atmospheric carbon is in the form of CO2, gas with a minor fraction present as gaseous CH4 and CO. CO2 is removed from the atmosphere by photosynthesis on land and in the ocean and via the chemical weathering of crustal rocks.[8] Most of the Earth's carbon—about 65 billion tonnes—is stored in rocks. The rest is in the oceans, the atmosphere, trees and plants, soil, and of course: fossil fuels."[9]
CC: "The slow-rate geochemical processes, including the formation and burial of carbonates, the burial of organic matter (on land or in the ocean), and volcanism have largely determined the flow of CO2 into and out of the atmosphere on multi-million-year time scales. As currently accepted, the reduction of atmospheric CO2 caused by these burial processes appears to be bound by tectonic processes, which return the carbon from the Earth’s mantle and crust to the atmosphere through volcanic degassing (which became less frequent as the Earth’s mantle progressively cooled). Later, the flow of atmospheric CO2 began to be controlled by other natural processes such as the origin and the expansion of forests, which caused increased burial of organic carbon by photosynthesis, and the respiration of living organisms, which added CO2 back into the atmosphere."[10][11][12]
Needs paraphrasing...
"As naturally occurring compounds, the carbon based gases CO2 and CH4 are part of the global carbon cycle that involves the movement of carbon from one reservoir to another via numerous pathways and processes, including fast and slow cycles. The slow cycle involves geological processes such as rock formation and weathering, and moves through the environment over millions of years. The fast carbon cycle involves the cycling of carbon through organic matter. Plants and phytoplankton uptake atmospheric carbon dioxide (CO2) in the process of photosynthesis, producing sugar used for energy and growth, and oxygen as a waste product. During the burning of carbohydrates for energy by microbes, plants, by animals which have eaten the plants (or animals which have eaten the animals which have eaten the plants), or by fire, CO2 is returned to the atmospheric pool. These cycles can be very short (hours-days) or last centuries and can be affected by external drivers such as solar energy inputs, orbital shifts, vegetation clearance, soil loss and fossil fuel burning."[13]
Needs paraphrasing...
"By contrast, the fast carbon cycle moves carbon relatively quickly through the atmosphere and biosphere, where it supports most of the life on Earth. Several billion tonnes of carbon may move around this cycle each year — much more than circulates through the slow cycle. Plants and phytoplankton are the main components of the fast carbon cycle. They absorb and consume atmospheric carbon dioxide through photosynthesis, which is then returned to the atmosphere when the plant or organism eventually dies and decays — or is destroyed by fire if it consumes the biomass."[9]
"But the largest flux in the fast carbon cycle is that produced by the combustion of fossil fuels — which produces copious amounts of carbon dioxide and other greenhouse gases which are all expelled into the atmosphere. Much of this carbon finds its way back to the land and into the oceans, but a significant fraction remains in the atmosphere where the concentration of carbon dioxide and methane continues to increase — directly contributing to global warming."[9]
Needs paraphrasing...
"Most of the carbon is stored in rocks, and the rest in the ocean, (dissolved inorganic carbon, living and non-living marine biota), atmosphere, soil, fossil fuels, and plants. The movements of carbon occur because of various chemical, physical, geological, and biological processes. Carbon flows between each reservoir in a biogeochemical loop known as the carbon cycle, which has slow and fast exchanges of components. Billions of tons of carbon in the form of CO2 are absorbed by oceans and living biomass known as sinks, and are emitted into the atmosphere through a natural process, called sources."[14]
"When in equilibrium, carbon fluxes between these reservoirs are roughly balanced and this balance helps to keep earth's temperature relatively stable—by trapping some of the outgoing energy the earth radiates back which it received from the sun and thus acting like the glass panels of a greenhouse. Thanks to greenhouse gases (water vapor, CO,, methane, nitrous oxide, and ozone), the earth's average temperature is 60°F more hospitable for living creatures. Without this natural greenhouse effect, temperatures would be lower than they are now, and life as it is known today would not be possible. Any change in the cycle, shifts carbon from one reservoir and places more carbon in the other reservoirs. The transformations which add carbon gases into the atmosphere lead to the warmer temperatures on earth. The earth's temperature is maintained through a slow carbon cycle, like a thermostat over several thousands of years. However, short-term temperature variations may occur as evidenced from swinging of earth between ice ages and interglacial periods. That is, in the slow carbon cycle, carbon takes nearly 1-200 million years to move between rocks, soil, ocean, and atmosphere — approximately 10-100 million metric tons (10^13-10^14 g) shift annually. While the fast carbon cycle moves 10^16-10^17 g every year, as human activities add 10^15 g."[14]
"The shift of carbon from the atmosphere to rocks (lithosphere) starts with rain, as the atmospheric carbon in the form of carbon dioxide combines with water to form carbonic acid (CO2 + + HC00). This weak acid dissolves the rock through a process called chemical weathering and releases calcium, potassium, sodium and magnesium ions. Flowing water bodies such as rivers transport these ions to the ocean. The calcium ions in the ocean com-bine with bicarbonate ions to form a chalky white substance called calcium carbonate, an active component in antacids. A small portion of calcium carbonate in the ocean is made by calcifying (shell building) marine organisms such as corals and marine plants such as coccolithophores. When these organisms die, they sink down to the sea floor and over time, the layers of shells and sediment am cemented together to form rooks and the carbon is stored in the form of stones, as lime stones and its derivatives. On our planet, only 80% of rocks are formed in this way. while the rest are composed of carbon from living organisms, namely organic carbon embedded in mud layers. Over millions of years, heat and pressure compress the mud and carbon to form shale-like sedimentary rocks. While the dead mater of plants build up faster than their decay, the organic carbon deposits become oil, coal, or natural gas. The surfaces of both land and ocean of the Earth are formed on dynamic platelets, and during their movements, when the platelets collide, one is placed over another and melts under extreme heat and pressure. During the eruption of volcanoes, the heated rocks recombine to form silicates, covering the land with fresh silicate rocks and releasing carbon dioxide into the atmosphere through as low cyclic process. Annually, volcanoes emit between 125 and 375 million metric tons of carbon dioxide, while human activities add nearly 30 billion tons (around 100-300 times more than volcanoes) of carbon dioxide per year. It is the chemical reactions which regulate the carbon flux between ocean, land, and atmosphere."[14]
"The carbon cycle is considered to be fast if the movement of carbon takes the time of a lifespan and occurs mainly within biosphere through living organisms. Carbon plays a key role in bio-systems due to its ability to form several bonds. leading to numerous varieties of complex organic molecules, and many organic molecules make up chains and rings through carbon atoms that have established a strong bond with other carbon atoms. As an example. DNA is composed of a couple of intertwined molecules built around a carbon chain. The bonds in the long carbon chains can store lots of energy, and hence when breaking them apart, the stored energy is released, thereby making carbon components an excellent source of fuel for living beings. Plants and phytoplankton (microscopic marine organisms) are the two key components of the fast carbon cycle, as they absorb atmospheric carbon dioxide into their cells. Using solar energy, they combine carbon dioxide and water to form sugar and oxygen. The carbon movement from plant to atmosphere involves four main steps: Plants break down the sugars to obtain energy for their growth, while animals including humans consume plants/plankton to break down the plant sugar to derive energy. When the plants and plankton die and decay (fire consuming the plants or being eaten by bacteria), oxygen combines with sugar to release water, carbon dioxide and energy. These chemical reactions are given below: CO, + H,0 + Energy CH, 0 + 0, ( 1.1) CH,0 + 0, •-• CO, + H, 0 + Energy (1.2) In all four processes, carbon dioxide is released finally into the atmosphere. The plant life on Earth is so closely knit together with the fast carbon cycle as can be clearly seen during winter in the northern hemisphere, only very few land plants grow and many decay, leading to of atmospheric carbon dioxide concentrations escalating. During spring, plants begin to grow again and a drop in atmospheric carbon dioxide concentration can be noted—as though the Earth is breathing."[14]
Needs paraphrasing...
"Photosynthesis creates new particulate organic matter (POM). An important consequence of photosynthesis has been the provision of O2 gas that degasses into the atmosphere. About half of the global O2 production is currently supported by marine photosynthesis. In the ocean, most of the carbon is in the form of inorganic molecules, i.e., bicarbonate ( HCO−
3), carbonate (CO2−
3), and carbon dioxide (CO2). These are collectively referred to as dissolved inorganic carbon (DIC). The vast majority of the DIC is bicarbonate. Most of the phosphorus is dissolved and in the form of orthophosphate. Most of the nitrogen is dissolved, with the organic form being nearly as abundant as the inorganic forms. The most abundant form of dissolved inorganic nitrogen is nitrate."[8]
- references
- ^ a b c Lürling, Miquel (2020). "Grazing resistance in phytoplankton". Hydrobiologia. 848: 237–249. doi:10.1007/s10750-020-04370-3. S2CID 221110997.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Falkowski, P.; Scholes, R. J.; Boyle, E.; Canadell, J.; Canfield, D.; Elser, J.; Gruber, N.; Hibbard, K.; et al. (2000). "The Global Carbon Cycle: A Test of Our Knowledge of Earth as a System". Science. 290 (5490): 291–296. Bibcode:2000Sci...290..291F. doi:10.1126/science.290.5490.291. PMID 11030643. S2CID 1779934.
- ^ Smith, T. M.; Cramer, W. P.; Dixon, R. K.; Leemans, R.; Neilson, R. P.; Solomon, A. M. (1993). "The global terrestrial carbon cycle". Water, Air, & Soil Pollution. 70 (1–4): 19–37. Bibcode:1993WASP...70...19S. doi:10.1007/BF01104986. S2CID 97265068.
- ^ Lade, Steven J.; Donges, Jonathan F.; Fetzer, Ingo; Anderies, John M.; Beer, Christian; Cornell, Sarah E.; Gasser, Thomas; Norberg, Jon; Richardson, Katherine; Rockström, Johan; Steffen, Will (2018). "Analytically tractable climate–carbon cycle feedbacks under 21st century anthropogenic forcing". Earth System Dynamics. 9 (2): 507–523. Bibcode:2018ESD.....9..507L. doi:10.5194/esd-9-507-2018.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Mills, Benjamin J.W.; Krause, Alexander J.; Scotese, Christopher R.; Hill, Daniel J.; Shields, Graham A.; Lenton, Timothy M. (2019). "Modelling the long-term carbon cycle, atmospheric CO2, and Earth surface temperature from late Neoproterozoic to present day". Gondwana Research. 67: 172–186. Bibcode:2019GondR..67..172M. doi:10.1016/j.gr.2018.12.001. S2CID 135192070.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Hain, M. P., Sigman, D. M., and Haug, G. H.: (2014). "The Biological Pump in the Past". In: Treatise on Geochemistry, Second Edition, Turekian, H. D. H. and Karl, K., Elsevier (Eds.), Oxford, 485–517.
- ^ Cartapanis, Olivier; Galbraith, Eric D.; Bianchi, Daniele; Jaccard, Samuel L. (2018). "Carbon burial in deep-sea sediment and implications for oceanic inventories of carbon and alkalinity over the last glacial cycle". Climate of the Past. 14 (11): 1819–1850. Bibcode:2018CliPa..14.1819C. doi:10.5194/cp-14-1819-2018. S2CID 56214252.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Libes, Susan M. (2015). Blue planet: The role of the oceans in nutrient cycling, maintain the atmosphere system, and modulating climate change In: Routledge Handbook of Ocean Resources and Management, Routledge, pages 89–107. ISBN 9781136294822.
- ^ a b c Cite error: The named reference
Bush2020was invoked but never defined (see the help page). - ^ Royer, D.L. (2014) "Atmospheric CO2 and O2 during the Phanerozoic: Tools, patterns, and impacts". In: Treatise on Geochemistry; Turekian, K., Holland, H. (Eds.), Elsevier Science, pp. 251–267
- ^ Bergman, N. M. (2004). "COPSE: A new model of biogeochemical cycling over Phanerozoic time". American Journal of Science. 304 (5): 397–437. Bibcode:2004AmJS..304..397B. doi:10.2475/ajs.304.5.397.
- ^ Cite error: The named reference
Carpinteri2019was invoked but never defined (see the help page). - ^ Kay, Martin (2019) "Impacts of warming, drought and sea level rise on ombrotrophic peatlands". Doctoral thesis, Manchester Metropolitan University.
- ^ a b c d Aulice Scibioh, M.; Viswanathan, B. (2 January 2018). Carbon Dioxide to Chemicals and Fuels. pp. 3–4. ISBN 9780444639974.
Particulate Inorganic Carbon (PIC)
[edit]Concentrations of PIC (i.e., calcium carbonate or calcite).
- carbonate compensation depth
- aragonite compensation depth
- lysocline
- calcareous ooze
- Carbonate pump
- snowline: the depth at which carbonate disappear from sediments under steady-state conditions
§§§ NEEDS PARAPHRASING...
"The oceans are full of plant life which provides food for all the larger animals in the oceans. This plant life is really small and you can only see individual plants with a microscope. However, when there is a lot of this plant life in one place, it can change the color of the ocean so much that we can see it from a ship, a plane or even from satellites. We call these plants algae, or phytoplankton. Just like on the land, where there are lots of different types of plant, there are lots of different types of phytoplankton. We are interested in one particular type, which has a chalk outer shell, causing the ocean to turn a milky blue when there are lots of them growing together. These chalk covered phytoplankton play a major role in regulating carbon in the oceans, and so it is important to know both where these phytoplankton are and how many of them there are. We have developed a new way to estimate how much chalk is in the ocean from satellite observations to help us estimate where these chalk covered phytoplankton are."[1]
Great calcite belt
[edit]
§§§ NEEDS PARAPHRASING...
"Particulate inorganic carbon (PIC), or calcium carbonate, is a major component of the global ocean carbon cycle. Through the process of calcification, marine organisms produce PIC shells and carbon dioxide from calcium ions and bicarbonate in seawater. These organisms, upon death, eventually sink to the ocean floor. During this process, the calcium carbonate shells may partially dissolve and what remains accounts for about 75% of carbon deposition on the seafloor (Groom & Holligan, 1987). Moreover, calcium carbonate production leads to an increase in partial pressure of dissolved carbon dioxide in the surface layer of the ocean, weakening the effectiveness of the carbon dioxide sink produced by photosynthesis (Shutler et al., 2013)."[1]
"There are many calcifying marine organisms, including coral, foraminifera, and pteropods, but coccolithophores are a major producer of PIC in the pelagic zone of oceanic regimes (Milliman, 1993). Coccolithophores produce intricate calcium carbonate platelets, known as coccoliths, which are extended around the cells (Paasche, 2002). During a bloom, the coccoliths can detach from the coccolithophores, becoming suspended in the water column."[1]
"Coccolithophores and their detached coccoliths can have a major impact on the oceanic light field due to their strong scattering nature. Typically, in nonbloom conditions 10–20% of the light backscattered from the ocean is due to detached coccoliths, whereas, in bloom conditions this can be greater than 90% (Balch et al., 1991, 1999). The strong scattering characteristics of coccolithophores and their associated PIC result in enhanced reflectance across the visible spectrum (400–700 nm)."[1]
"From the dawn of satellite ocean color measurements, large coccolithophore blooms have been visible as highly reflective regions in satellite imagery (Holligan et al., 1983). Gordon et al. (2001) and Balch et al. (2005) developed algorithms to estimate the PIC concentration of the surface layer of the water column from water‐leaving radiance. Both of these methods use absolute values of water‐leaving radiances rather than radiance ratios which are typically used when estimating chlorophyll‐a, because PIC increases the radiance uniformly in the blue and green (Gordon et al., 1988). Currently, an algorithm merging the approach of Gordon et al. (2001) and Balch et al. (2005) is included as a standard product in the data distributed by NASA's Ocean Biology Processing Group."[1]
Overview
[edit]- FROM: Total inorganic carbon:
Question 8. How will lower calcification rates, due to an increase in ocean acidification, higher ocean temperatures, and changes in nutrients affect ocean carbon chemistry and carbon export rates? Answer. Acidification will tend to reduce the calcification in the upper ocean, the sinking flux (export) of particulate inorganic carbon, and the remineralization of particulate inorganic in the subsurface ocean. The effect of acidification on total biological productivity in the surface ocean may be about neutral, as it is likely that non-calcifying organisms may be able to replace calcifying phytoplankton populations that are diminished due to acidification. Organic carbon export to the subsurface ocean via sinking particles is not directly proportional to biological productivity, but depends upon the composition of the food web. Organic matter has a density similar to seawater, and there is evidence indicating that heavier ballast materials, such as carbonate shells, increase organic matter sinking rates. The impact of reduced calcification on the export of organic carbon in the open ocean is less certain, but may also result in a reduction in export... Reduced inorganic export has the opposite effect as reduced organic carbon export on surface water chemistry and air-sea carbon fluxes. The formation of organic matter lowers seawater dissolved inorganic carbon (DIC) and lowers the partial pressure of carbon dioxide (pCO2), which governs the air-sea gas exchange of carbon dioxide. A reduction in organic matter export, therefore, would reduce the effectiveness of the biological pump and act to increase surface water and atmospheric CO2 thus accelerating climate change. The formation of calcium carbonate (CaCO3) or calcification in surface waters lowers both seawater DIC and alkalinity (a measure of the acid-base balance of seawater). For each mole of CaCO3 removed, DIC drops by 1 mole and alkalinity drops by 2 moles. Somewhat counter intuitively, calcification increases pCO2 because the effect of the alkalinity change outweighs that of DIC. Therefore reduced carbonate export would act to decrease surface water and atmospheric CO2 thus helping to ameliorate climate change. Preliminary model simulations, however, suggest that the calcification-alkalinity feedback mechanism provides only a small brake on increasing atmospheric carbon dioxide due to fossil fuel combustion.[5]
- FROM: [4]
NEEDS PARAPHRASING "Particulate Inorganic Carbon Observations "The cycling of particulate inorganic carbon (i.e. CaCO3) in the ocean also affects the biological pump and therefore atmospheric CO2, but by more indirect mechanisms. Whether carbonates precipitate or dissolve can be directly linked to the saturation state (Ω) of the ocean (readers are referred to Box 1 for a brief primer on carbonate thermodynamics). Compared to the 5% of POC that is 620 exported from the euphotic zone and reaching the sediments, a significantly higher amount of PIC is vertically transported to the bottom of the ocean (about 50% of the PIC export flux, ?). The role of sinking particulate inorganic carbon in the biological pump is complex, because the deep dissolution of PIC is largely controlled by the degradation of sinking POC and releases alkalinity, which in turn titrates part of the CO2 released during POC degradation. In addition its high specific gravity plays 625 a key role for the sinking rates of biogenic aggregates (the ballasting effect) and thus the residence time of particulate carbon in the ocean (e.g., ??). The mechanisms responsible for carbonate dissolution in the ocean are still matter of debate (?). Global observations showing that the depth of the lysocline coincides with the saturation horizon (e.g. ?) have been used to imply that thermodynamic constraints are a dominant control on calcium carbonate preservation. However, a kinetic control on 630 carbonate dissolution has been highlighted by in-situ experiments in the North Pacific (e.g., ?) and laboratory studies reveal that dissolution rates increase in undersaturated waters (??). In addition, observational evidence even points to a partial dissolution of sinking carbonate above the saturation horizon (e.g. ?). ? estimates, using global production estimates of CaCO3 and globally averaged deep water sediment trap data, that probably 40–80% of the calcium carbonate produced in the sur635 face ocean dissolves in the upper water column. However, the mechanisms that drive the dissolution of carbonates above the lysocline remain enigmatic. The dissolution of carbonates within acidic micro-environments, such as the digestive system of zooplankton or marine aggregates have been..."

Biogenic CaCO3 tests are produced in the photic zone of the oceans (green circles). Upon death, those tests escaping dissolution near the surface, settle along with clays materials. Above the saturation horizon, waters are supersaturated and CaCO3 tests are largely preserved. Below the saturation, waters are undersaturated because of increasing solubility with depth and the release of CO2 from organic matter decay and CaCO3 will dissolve. Dissolution occurs primarily at the sediment surface as the sinking velocity of debris is rapid (broad white arrows). At the carbonate compensation depth, the rate of dissolution exactly matches rate of supply of CaCO3 from above. At steady state this carbonate compensation depth is similar to the snowline (the first depth where carbonate poor sediments occur). The lysocline is the depth interval between the saturation and carbonate compensation depth.[7][6]
- references
- ^ a b c d e Mitchell, C.; Hu, C.; Bowler, B.; Drapeau, D.; Balch, W. M. (2017). "Estimating Particulate Inorganic Carbon Concentrations of the Global Ocean from Ocean Color Measurements Using a Reflectance Difference Approach". Journal of Geophysical Research: Oceans. 122 (11): 8707–8720. Bibcode:2017JGRC..122.8707M. doi:10.1002/2017JC013146.
- ^ Kandasamy, Selvaraj; Nagender Nath, Bejugam (2016). "Perspectives on the Terrestrial Organic Matter Transport and Burial along the Land-Deep Sea Continuum: Caveats in Our Understanding of Biogeochemical Processes and Future Needs". Frontiers in Marine Science. 3. doi:10.3389/fmars.2016.00259. S2CID 30408500.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Bauer, James E.; Cai, Wei-Jun; Raymond, Peter A.; Bianchi, Thomas S.; Hopkinson, Charles S.; Regnier, Pierre A. G. (2013). "The changing carbon cycle of the coastal ocean". Nature. 504 (7478): 61–70. doi:10.1038/nature12857. PMID 24305149. S2CID 4399374.
- ^ Regnier, Pierre; Friedlingstein, Pierre; Ciais, Philippe; MacKenzie, Fred T.; Gruber, Nicolas; Janssens, Ivan A.; Laruelle, Goulven G.; Lauerwald, Ronny; Luyssaert, Sebastiaan; Andersson, Andreas J.; Arndt, Sandra; Arnosti, Carol; Borges, Alberto V.; Dale, Andrew W.; Gallego-Sala, Angela; Goddéris, Yves; Goossens, Nicolas; Hartmann, Jens; Heinze, Christoph; Ilyina, Tatiana; Joos, Fortunat; Larowe, Douglas E.; Leifeld, Jens; Meysman, Filip J. R.; Munhoven, Guy; Raymond, Peter A.; Spahni, Renato; Suntharalingam, Parvadha; Thullner, Martin (2013). "Anthropogenic perturbation of the carbon fluxes from land to ocean". Nature Geoscience. 6 (8): 597–607. Bibcode:2013NatGe...6..597R. doi:10.1038/ngeo1830. hdl:2268/150126.
- ^ Effects of climate change and ocean acidification on living marine organisms Hearing before the United States Senate, 10 May 2007.
 This article incorporates text from this source, which is in the public domain.
This article incorporates text from this source, which is in the public domain.
- ^ a b Middelburg, Jack J. (2019). "Biogeochemical Processes and Inorganic Carbon Dynamics". Marine Carbon Biogeochemistry. SpringerBriefs in Earth System Sciences. pp. 77–105. doi:10.1007/978-3-030-10822-9_5. ISBN 978-3-030-10821-2. S2CID 104368944.
- ^ Boudreau, Bernard P.; Middelburg, Jack J.; Luo, Yiming (2018). "The role of calcification in carbonate compensation". Nature Geoscience. 11 (12): 894–900. Bibcode:2018NatGe..11..894B. doi:10.1038/s41561-018-0259-5. S2CID 135284130.
Carbon forms
[edit]
(b) Carbon forms include dissolved inorganic carbon (DIC), organic matter (OM), particulate organic matter (POM), dissolved organic matter (DOM), and particulate inorganic matter (PIC).
| External videos | |
|---|---|
Phytoplankton blooms
[edit]
- Algal blooms
- Diatom blooms
- Coccolithophore blooms
- Cyanobacteria blooms
- Dinoflagellate blooms
- Ciliate blooms: Mesodinium rubrum is a "globally distributed nontoxic ciliate that is known to produce intense red-colored blooms using enslaved chloroplasts from its algal prey".
Light attenuation
[edit]
Miscellaneous
[edit]

- marine bacterioplankton
"The microbes that make up the "ocean constellations" represent the some of the Earth’s most abundant organisms: marine bacterioplankton. There are several hundred thousand to one million bacteria in every drop (one milliliter) of ocean water. When scaled to the total volume of the ocean (i.e. ~ 1.5 billion km3), there are approximately 120,000,000,000,000,000,000,000,000,000 (1.2 x 1029) bacterioplankton cells in the global ocean, or ~ 95% of the all the living biomass in the sea. In other words, the total marine bacterioplankton biomass is greater than all of the ocean’s zooplankton, shellfish, fish and whales summed together."[3]
"The majority of this unseen marine bacterioplankton grow very fast, dividing every couple of days. To meet their metabolic demands, bacterioplankton must consume vast amounts of organic compounds like carbohydrates and proteins and recycle inorganic nutrients like carbon dioxide, nitrate and phosphate every day. Though each of these microbes live on the timescales of days and work on spatial scales of nanometers (a billionth of a meter), their sheer numbers and high growth rates mean they affect ocean chemistry on the scales of ecosystems. These marine microbes are the true drivers of large scale biogeochemical cycles in oceanic systems as we are witnessing in the subarctic Pacific Ocean."[3]
"With EXPORTS, we are examining the intricate mechanisms of the oceanic biological “carbon pump”. The carbon pump describes how photosynthically fixed carbon (usually from atmospheric carbon dioxide that has dissolved in the ocean) is processed and reprocessed through the ocean’s planktonic food web in the surface layers of the ocean. Ultimately a portion of the carbon is exported out of the suface by sinking and mixing to the deep ocean hundreds to thousands of meters below.[3]
"How exactly the ocean absorbs and retains carbon largely depends in part on the growth of microbes but also where in the water column (between the ocean surface and abyss) they intercept and consume sinking organic particles or dissolved organic matter. For carbon, and other associated elements, to be stored for long periods of time in the ocean, it first must travese the portion of the ocean’s water column that extends between ~one hundred and one thousand meter depths. This zone is referred to by oceanographers as the ocean’s twilight zone. The twilight zone is so named not only because there is little to no light but also because the chemical budgets and biological processes that occur in this zone are not well understood. If microbes recycle the organic matter too close to the surface, the carbon escapes long-term storage. But if the sinking or mixed organic matter gets deep enough, then that carbon is removed from interaction with the atmosphere for decades to millennia.[3]
"Our project is designed to investigate marine microbes and their interaction with organic compounds throughout the water column of the subarctic Pacific Ocean. We are conducting experiments at sea that combine tools from ecology, molecular biology, and marine chemistry to investigate how bacterioplankton consume the organic substrates available to them. By examining microbial behavior, we will learn more about how the ocean’s “inner space” functions and how biology helps govern the movement of carbon within Earth’s systems.[3]
Dwindling diatoms
[edit]
Viral loop
[edit]



in Amazonian lakes [6]
Mixotrophs
[edit]- Mixotrophic protists and a new paradigm for marine ecology: where does plankton research go now? 2019 not CC by
- * The evolution of microbial ecology of the ocean 2018 not CC by <<<<< —————— <<<<<
- Bridging the Gap between Knowing and Modeling Viruses in Marine Systems—An Upcoming Frontier 2017 - *** CC by
- Dynamic Ecology - an introduction to the art of simulating trophic dynamics. – Kevin Flynn
- Incorporating mixotrophy into mainstream marine research. – mixotroph.org
- Mixotrophy everywhere on land and in water: the grand écart hypothesis 2016 - not CC by
- Mixotrophy in Microorganisms: Ecological and Cytophysiological Aspects 2013 - not CC by
- The mixotrophic nature of photosynthetic plants 2013 - not CC by
- Weaving marine food webs from end to end under global change 2011 - not CC
Cited by 177: Mixotrophy in the Marine Plankton 2017[7]
Mixotrophy in Protists: From Model Systems to Mathematical Models 2018[8]

"If (mixotrophs) weren’t in the oceans, we’re suggesting atmospheric carbon dioxide might be higher, because there would less of the large, carbon rich particles formed which efficiently transfer carbon to depth," Mick Follows says.[26]




- Mixotrophy
Aquatic protists are often categorized as either strict heterotrophs or autotrophs, despite increasing evidence that mixotrophs are the default and strict heterotrophs/autotrophs are the exception.[33] Mixotrophs are protists that use both phototrophy and phagotrophy to obtain energy and increase their biomass.[34] A majority of protist groups, with the exception of diatoms, express some level of mixotrophy between a gradient of strict heterotrophy and strict autotrophy, with most being primarily a phagotroph or a phototroph that supplements their growth with alternative energy sources.[33][29]
Grazing in the protist community is typically estimated indirectly through prey removal experiments [35] or the dilution method.[36] Grazers in these types of experiments are identified by their lack of chloroplasts, while mixotrophs have to be identified as protists that contain both chloroplasts and prey.[37] Therefore, grazing by mixotrophs has to be measured directly through the uptake of fluorescently labeled bacteria,[38] fluorescently labeled algae,[39] or fluorescent microspheres.[40] Thus, unless a study is specifically focused on quantifying grazing or ingestion rates by mixotrophs, all grazing rates measured by standard methods are usually attributed to pigmentless protists (strict heterotrophs).[29]
The miscategorization of mixotrophs as strict autotrophs or heterotrophs has major implications for the way the flow of carbon and nutrients through the planktonic food web is understood (see above diagram). Mixotrophs can increase trophic transfer efficiency by offsetting some of the carbon lost to respiration in secondary and tertiary plankton producers with carbon from photosynthesis.[28][29]
Of the cryptic interactions presented here, mixotrophy is the most researched. Mixotrophy has been studied for decades (Stoecker 1998),[34] modeled in a variety of ways (Stickney et al. 2000,[41] Flynn and Mitra 2009,[42] Ward et al. 2011),[43] and incorporated into recent food web models (D’Alelio et al. 2016,[44] and Ward and Follows 2016[28]). However, while mixotrophs have been included in recent food web models, a majority of new models do not account for it and the ecological data required to accurately model this interaction and ground truth the results are scarce. Currently, a generalized understanding of the relative occurrence of mixotrophs compared to strict autotrophs and heterotrophs is unknown because most studies of mixotrophy focus on the grazing impact of a particular species or groups of protists.[45] This approach only provides an estimation of the impact of mixotrophy during blooms of particular species.[29]
A few studies conducted in a small range of specific environments have determined the grazing impact that mixotrophs can have on the consumption of bacteria compared to strict heterotrophs. These studies have shown that the consumption of bacteria by mixotrophs can be less than,[46] equal to,[47] or even more than heterotrophs (Dominique et al. 2003),[48] depending upon the time of year and study location. However, in order to improve recently developed food web models that account for mixotrophs, more data on variability of and grazing impact of mixotrophic assemblages in the environment is needed.[29]
There are numerous examples of mixotrophs being modeled. Stickney et al. (2000) expanded on the basic nutrient– phytoplankton–zooplankton–detritus model to include a node for mixotrophy. Flynn and Mitra (2009) created a complex mechanistic model that described how mixotrophs interacted with carbon, nitrogen, and phosphorus. Ward et al. (2011) developed an idealized model that modeled the physiology of different plankton to determine which environmental conditions favored mixotrophs. D’Alelio et al. (2016) produced a mass balance model that computed biomass flow between 63 functional nodes, ten of which were mixotrophic groups or species. Ward and Follows (2016) designed a simplified global model of the plankton foodweb that removed phytoplankton and zooplankton, modelling plankton as mixotrophs on a spectrum between strict autotrophy and heterotrophy. However, the models by Stickney, Flynn and Mitra, and Ward are examples of modelling mixotrophs rather than incorporating mixotrophs into complex food web models. The Ward and Follows model provides an estimation of the role mixotrophs have in the transfer of carbon, but it is theoretical and still needs to be validated with field observations. The D’Alelio model incorporates mixotrophs, uses some real data, and accounts for the different trophic strategies mixotrophs may have under different conditions.[29]
Ward and Follows (2016) developed a theoretical model that compared the transfer of carbon within a food web with strict autotrophs and heterotrophs to a food web with only mixotrophs. This model did not require any detailed quantitative data, but produced results that could be groundtruthed by the current understanding of the food web. Importantly, this study highlighted the types of data collection on mixotrophs that were further needed, specifically a more generalized understanding of the portion of mixotrophs within each plankton size group. Studies like Ward and Follows (2016) can help direct the focus of research done by ecologists and provide testable hypotheses for their projects.[29]
-
Venus flytrap photosynthesing and eating insects[30]
-
Mixotrophic phytoplankton Karlodinium capturing and ingesting a small cell[30]
-
The mixotroph Dinophysis feeding on Mesodinium. The small red circles inside the Mesodinium are algae the Mesodinium has already eaten. The arrow points to the small straw or feeding tube it uses to feed on Mesodinium.[30]

- Introduction
"The traditional view of the planktonic fond web is simplistic: nutrients are consumed by phytoplankton that, in turn, support zooplankton that ultimately support fish. Historically. harmful algal species have been viewed as phototrophic phytoplankton. although with come notable exceptions. This primary producer-centric structure is the foundation of most models that have been used to explore fisheries production, biogeochemical cycling, and climate change. In recent years. however, the importance of mixotrophs increasingly has been recognised, changing scientists' view of these traditional food web interactions Katechakis et al. 2005: Glibert and Legrand 2006; Burkholder et al. 2008; Jeong et al. 2010; Hansen 2011: Flynn et al. 2013; Mite et al. 2016." [50]
"Mixotrophy, the combination of phototrophy and heterotrophy (phago- and/or osmotrophy), enables planktonic protists traditionally labelled as "phytoplankton" or "microzooplankton" to function at multiple trophic levels. Harmful algal blooms (HABs), including both high-biomass bloom formers and toxic species, are now recognised to be not only more prevalent in waters worldwide but also typically mixotrophic (Burkholder et al. 2008). Thus, it is important that concerted efforts are taken to understand mixotrophy as an important nutritional strategy that alters food web dynamics."[50]
"The processes of mixotrophy can radically change the dynamics and efficiency of activities of the "phytoplankton" and "microzooplankton" (Flynn ad Mitre 2009; Mita and Flynn 2010; Flynn et al. 2013; Mitra et al. 2014b). Mixotrophy includes the concept of osmotrophy, the uptake of organic substrates, and phagotrophy, the ingestion of particulate material, by those organisms normally though to be dependent on inorganic carbon (C) via photosynthesis. Osmotrophy is ubiquitous among the protists (e.g., Berman and Bronk 2003; Glibert and Legrand 2006); phagotrophy is also much more common than previously recognized and has direct implications for trophic dynamics (namely, grazing). A fundamental revision of the functional taxonomy of aquatic protists has recently been proposed in recognition of the importance of mixotrophy (Mitra et al. 2016)."[50]
"In this chapter we narrowly define mixotrophy m phoffitrophy with phagorrophy. We emphasize that mixotrophy warrants realistic descriptions within food web models; it repre-sents a more flexible strategy in nutrient acquisition, grovAh, and production, with far-reaching consequencm not only for growth of the mixotrophs but also for their competitors and prey. In essence, miffirophic organisms alter the flaw of nutrients through the planktonic system, potentially upgrading it biochemically and changing the nature of predator-prey interactions in a highly efficient manner. An apprecia-tion of the importance of mixotrophy also affects which ,orient for. Mould be considered in (ewer management strategies; for example, if mixonophic HABs are promoted by events such m bacterial gmvAls on organic substances, through ingestion of bacteria then dissolved organks as well as the usual dissolval inor-ganic nutrient loads require consideration" [50]
"... In the ensuing decades, it has been recognized that mixotrophic organisms. many of which include HAB species (e.g., Burkholder et al. 2008). are nor only much more common than originally thought, but also their physiology is now known to be far more complex than could be described by simply bolting ingether descriptions of the contrasting processes of phototrophy and phagotrophy (Mitre and Flynn 2010)..."[50]
- Mixotrophy across the Spectrum of Nutrient Supply
"Planktonic mixotrophy has long been considered important in oligotrophic habitats. where piaentially limiting elements am much more concentrated in microbial prey Ilan in the water column Hones Prnet. More recently it has been recognised that mixotiophy is also important in marophic eAtallirn and coastal marine embayments, where mar, OAR taxa are mixotrophic (Burkholder et al. ESN. In such environ-ments, mixtgrophs respond both directly to nutrient inputs and indirectly through high abundance of Iwterial and algal prey Mat are stimulated by the elevated nutrients. It is wortb noting Mat M aa-called marophic systems, light c. often be .d hence limit newly fixed C. Gass, mixotrophy provides a mechanism to exploit alternative routes for the acquisition of C. Another important facet of mixatrophy is that all mixmrophs are not Me same; Mitre et al. 001 61 divide these prteists into those that have a constitutive capacity for mixotmphy and Mom that acquire phototrophy by acquisition of photasystems from other organisms. The conaitutive mixotrophs include HAB species such m toxigenic taxa (e.g., the dinollagellate Sur(refiniunt and the haptophyte Prymnesium), as well as on-HAB species such as the large dinollagellate Cerium."[50]
"Among the non-constitutive mixotrophs, some, the plwaidic ciliatre, are generalists and can steal chlomplasts from a range of prey types, while others are specialists making use of plastids acquired from very few prey murces. These include the nontoxic "red tide. ciliate AlesmlininmAlyrionerra and the ioxigenic dinoflagellme DimmAysis, which actually obtains its plastids tard hand.' from Mesmlinium (Hansen et al. 2013 and references therein). Another group of nonmonstinnives that harbours symbiotic phototrophs includes the biogeochemically important foraminifera and radiolaria and also the HAB-fonning Mnoflagellate, "green" Pomes et el. 2014; Stoecker et B. 2017; see also Chap. 17, Goes el A. (2012)t 'Mese contrasting mixotrophie snategies impart different ecological impacts through the need and removal of prey (peMaps competitors) that could, in turn, Brea the operation of other trophic pathways. Mixotmphic HAfts may thus be expected to M msociatod with complex ecological scenarios. Many different types of harmful microalgal species thrive in eutrophying or eutrophic estuarine and coastal marine habitats, including cyanobemeria. dim-flagellates, haptophyte flagellates, hetendcontophyres (diatom, golden flagellates, brown-tide algae, and raphidophycean flagellates) (Burkholder et al. 20011, Graham et at 2016; Fig. 7.1). In 00, the bulk of eukaryotre planktonic purists commonly classed as phmotrophs is actually mismrophic (Flynn et 2011: Mimiet al. 20160 Thus, it appears that most phyreflagellates (heterokontoplimes, haptopbytes. 00 cryplophyles) are capable of consuming bacteria and small eukaryolic ce10 (Unrein et al. 2002; Pubkov and Tartan 2008; Stukel et at 2011)."[50]
"Dinollagellates exhibit a wide range in ,abilities, from Mose that express phagotrophy only upon extreme nutrient starvation to exclusively phago.phic forms. Even the latter organisms may gain at least transitory inorganic C-fixing capacity from ingestion of phototrophs (Raven M al. 2000: -Hong el at 2010: Hansen 2011) Of other mixarophs (Minnhagen et al. 2011). Some potentially toxic s,cies appear to have relatively poor affinities thigh Ks values) for ifi0ke of imtganic N and P substrates (DIN. dissolved inorganic N. DIP, dissolved inorganic IPh and may therefore depend on mixotrophy as a means to supplement their acquisition of dissolved substrates (Smay0 19977 A contrary argument is that because of mixotrophy (phagourphy being Me ancestral stale in pnatists—Mitre ei al. 2016), these species Md not need to evolve inorganic nutrient .nsponers with high affinity. Imponantly. mixotrophy also may permit growth to be sustained or even accelerated during periods of apparent water-column nutrient deficiency or imbalance (or real nutrient deficiency for norymixotmphic competi-tors) when only inorganic forms of dissolved nutrients are considered. The dine-flagellates Cochlodinium (Margaiffidinim) colArikoides and Karenia /wet, exemplify these advantages of rnixotrophy. When growing 0 a phototreph. C. polykrikoides has a growth rate of 0.17 dry day- Ileong et al. 2)60) Yet. when growing as a mixotroph, consuming cryptophyles, the division rate of... "[50]
See also
[edit]References
[edit]- ^ Fennel, K.; Alin, S.; Barbero, L.; Evans, W.; Bourgeois, T.; Cooley, S.; Dunne, J.; Feely, R. A.; Hernandez-Ayon, J. M.; Hu, C.; Hu, Xinping; Lohrenz, S.; Muller-Karger, F.; Najjar, R.; Robbins, L.; Russell, J.; Shadwick, E.; Siedlecki, S.; Steiner, N.; Turk, D.; Vlahos, P.; Wang, Z. A. (2018). Cavallaro, N; Shrestha, G; Birdsey, R; Mayes, M. A; Najjar, R; Reed, S; Romero-Lankao, P; Zhu, Z (eds.). "Chapter 16: Coastal Ocean and Continental Shelves. Second State of the Carbon Cycle Report". doi:10.7930/SOCCR2.2018.Ch16. S2CID 134528444.
{{cite journal}}: Cite journal requires|journal=(help) [1] - ^ Ocean Explorer NOAA. Updated: 26 August 2010.
- ^ a b c d e f The Inner Space of the Subarctic Pacific Ocean NASA Earth Expeditions, 4 September 2018.
 This article incorporates text from this source, which is in the public domain.
This article incorporates text from this source, which is in the public domain.
- ^ Rousseaux, Cecile S.; Gregg, Watson W. (2015). "Recent decadal trends in global phytoplankton composition". Global Biogeochemical Cycles. 29 (10): 1674–1688. Bibcode:2015GBioC..29.1674R. doi:10.1002/2015GB005139. S2CID 130686652.
- ^ a b c Mateus, Marcos D. (2017). "Bridging the Gap between Knowing and Modeling Viruses in Marine Systems—An Upcoming Frontier". Frontiers in Marine Science. 3. doi:10.3389/fmars.2016.00284. S2CID 21502988.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Almeida, Rafael M.; Roland, Fábio; Cardoso, Simone J.; Farjalla, VinãCius F.; Bozelli, Reinaldo L.; Barros, Nathan O. (2015). "Viruses and bacteria in floodplain lakes along a major Amazon tributary respond to distance to the Amazon River". Frontiers in Microbiology. 6: 158. doi:10.3389/fmicb.2015.00158. PMC 4349158. PMID 25788895.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Stoecker, Diane K.; Hansen, Per Juel; Caron, David A.; Mitra, Aditee (2017). "Mixotrophy in the Marine Plankton". Annual Review of Marine Science. 9: 311–335. doi:10.1146/annurev-marine-010816-060617. PMID 27483121.
- ^ Johnson, Matthew D.; Moeller, Holly V. (2018). "Editorial: Mixotrophy in Protists: From Model Systems to Mathematical Models". Frontiers in Marine Science. 5. doi:10.3389/fmars.2018.00490. S2CID 56482514.
- ^ Stoecker, Diane K.; Lavrentyev, Peter J. (2018). "Mixotrophic Plankton in the Polar Seas: A Pan-Arctic Review". Frontiers in Marine Science. 5. doi:10.3389/fmars.2018.00292. S2CID 52058871.
- ^ Ward, Ben A. (2019). "Mixotroph ecology: More than the sum of its parts". Proceedings of the National Academy of Sciences. 116 (13): 5846–5848. doi:10.1073/pnas.1902106116. PMC 6442617. PMID 30862733.
- ^ Mitra, Aditee; Flynn, Kevin J.; Tillmann, Urban; Raven, John A.; Caron, David; Stoecker, Diane K.; Not, Fabrice; Hansen, Per J.; Hallegraeff, Gustaaf; Sanders, Robert; Wilken, Susanne; McManus, George; Johnson, Mathew; Pitta, Paraskevi; Våge, Selina; Berge, Terje; Calbet, Albert; Thingstad, Frede; Jeong, Hae Jin; Burkholder, Joann; Glibert, Patricia M.; Granéli, Edna; Lundgren, Veronica (2016). "Defining Planktonic Protist Functional Groups on Mechanisms for Energy and Nutrient Acquisition: Incorporation of Diverse Mixotrophic Strategies". Protist. 167 (2): 106–120. doi:10.1016/j.protis.2016.01.003. PMID 26927496.
- ^ d'Alelio, Domenico; Eveillard, Damien; Coles, Victoria J.; Caputi, Luigi; Ribera d'Alcalà, Maurizio; Iudicone, Daniele (2019). "Modelling the complexity of plankton communities exploiting omics potential: From present challenges to an integrative pipeline". Current Opinion in Systems Biology. 13: 68–74. doi:10.1016/j.coisb.2018.10.003. S2CID 133896807.
- ^ Verma, Arjun; Barua, Abanti; Ruvindy, Rendy; Savela, Henna; Ajani, Penelope A.; Murray, Shauna A. (2019). "The Genetic Basis of Toxin Biosynthesis in Dinoflagellates". Microorganisms. 7 (8): 222. doi:10.3390/microorganisms7080222. PMC 6722697. PMID 31362398.
- ^ Stickney, H.L.; Hood, R.R.; Stoecker, D.K. (2000). "The impact of mixotrophy on planktonic marine ecosystems". Ecological Modelling. 125 (2–3): 203–230. doi:10.1016/S0304-3800(99)00181-7.
- ^ Flynn, Kevin J.; Mitra, Aditee; Anestis, Konstantinos; Anschütz, Anna A.; Calbet, Albert; Ferreira, Guilherme Duarte; Gypens, Nathalie; Hansen, Per J.; John, Uwe; Martin, Jon Lapeyra; Mansour, Joost S.; Maselli, Maira; Medić, Nikola; Norlin, Andreas; Not, Fabrice; Pitta, Paraskevi; Romano, Filomena; Saiz, Enric; Schneider, Lisa K.; Stolte, Willem; Traboni, Claudia (2019). "Mixotrophic protists and a new paradigm for marine ecology: Where does plankton research go now?". Journal of Plankton Research. 41 (4): 375–391. doi:10.1093/plankt/fbz026.
- ^ Selosse, Marc-André; Charpin, Marie; Not, Fabrice (2017). "Mixotrophy everywhere on land and in water: Thegrand écarthypothesis". Ecology Letters. 20 (2): 246–263. doi:10.1111/ele.12714. PMID 28032461.
- ^ Bjorbækmo, Marit F. Markussen; Evenstad, Andreas; Røsæg, Line Lieblein; Krabberød, Anders K.; Logares, Ramiro (2020). "The planktonic protist interactome: Where do we stand after a century of research?". The ISME Journal. 14 (2): 544–559. doi:10.1038/s41396-019-0542-5. PMC 6976576. PMID 31685936.
- ^ Berge, Terje; Chakraborty, Subhendu; Hansen, Per Juel; Andersen, Ken H. (2017). "Modeling succession of key resource-harvesting traits of mixotrophic plankton". The ISME Journal. 11 (1): 212–223. doi:10.1038/ismej.2016.92. PMC 5315484. PMID 27482925. S2CID 90620.
- ^ Mitra, Aditee; Castellani, Claudia; Gentleman, Wendy C.; Jónasdóttir, Sigrún H.; Flynn, Kevin J.; Bode, Antonio; Halsband, Claudia; Kuhn, Penelope; Licandro, Priscilla; Agersted, Mette D.; Calbet, Albert; Lindeque, Penelope K.; Koppelmann, Rolf; Møller, Eva F.; Gislason, Astthor; Nielsen, Torkel Gissel; St. John, Michael (2014). "Bridging the gap between marine biogeochemical and fisheries sciences; configuring the zooplankton link". Progress in Oceanography. 129: 176–199. doi:10.1016/j.pocean.2014.04.025.
- ^ Neo, Mei Lin; Eckman, William; Vicentuan, Kareen; Teo, Serena L.-M.; Todd, Peter A. (2015). "The ecological significance of giant clams in coral reef ecosystems". Biological Conservation. 181: 111–123. doi:10.1016/j.biocon.2014.11.004.
- ^ Livingstone, Paul G.; Morphew, Russell M.; Whitworth, David E. (2017). "Myxobacteria Are Able to Prey Broadly upon Clinically-Relevant Pathogens, Exhibiting a Prey Range Which Cannot be Explained by Phylogeny". Frontiers in Microbiology. 8: 1593. doi:10.3389/fmicb.2017.01593. PMC 5572228. PMID 28878752.
- ^ Jassey, Vincent E. J.; Signarbieux, Constant; Hättenschwiler, Stephan; Bragazza, Luca; Buttler, Alexandre; Delarue, Frédéric; Fournier, Bertrand; Gilbert, Daniel; Laggoun-Défarge, Fatima; Lara, Enrique; t. e. Mills, Robert; Mitchell, Edward A. D.; Payne, Richard J.; Robroek, Bjorn J. M. (2015). "An unexpected role for mixotrophs in the response of peatland carbon cycling to climate warming". Scientific Reports. 5: 16931. Bibcode:2015NatSR...516931J. doi:10.1038/srep16931. PMC 4658499. PMID 26603894.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Worden, A. Z.; Follows, M. J.; Giovannoni, S. J.; Wilken, S.; Zimmerman, A. E.; Keeling, P. J. (2015). "Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes". Science. 347 (6223). doi:10.1126/science.1257594. PMID 25678667. S2CID 206560125.
- ^ Gilbert, D.; Mitchell, E.A.D. (2006). "Chapter 13 Microbial diversity in Sphagnum peatlands". Peatlands - Evolution and Records of Environmental and Climate Changes. Developments in Earth Surface Processes. Vol. 9. pp. 287–318. doi:10.1016/S0928-2025(06)09013-4. ISBN 9780444528834.
- ^ Lindo, Zoë; Nilsson, Marie-Charlotte; Gundale, Michael J. (2013). "Bryophyte-cyanobacteria associations as regulators of the northern latitude carbon balance in response to global change". Global Change Biology. 19 (7): 2022–2035. Bibcode:2013GCBio..19.2022L. doi:10.1111/gcb.12175. PMID 23505142. S2CID 20493073.
- ^ Living a “mixotrophic” lifestyle MIT News, 1 February 2016.
- ^ Worden, A. Z., M. J. Follows, S. J. Giovannoni, S. Wilken, A. E. Zimmerman, and P. J. Keeling (2015) "Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes". Science, 347: 1257594. doi:10.1126/science.125.
- ^ a b c Ward, Ben A.; Follows, Michael J. (2016). "Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux". Proceedings of the National Academy of Sciences. 113 (11): 2958–2963. doi:10.1073/pnas.1517118113. PMC 4801304. PMID 26831076.
- ^ a b c d e f g h Millette, Nicole C.; Grosse, Julia; Johnson, Winifred M.; Jungbluth, Michelle J.; Suter, Elizabeth A. (2018). "Hidden in plain sight: The importance of cryptic interactions in marine plankton". Limnology and Oceanography Letters. 3 (4): 341–356. doi:10.1002/lol2.10084. S2CID 89941430.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c d e Glibert, Patricia M.; Mitra, Aditee; Flynn, Kevin J.; Hansen, Per Juel; Jeong, Hae Jin; Stoecker, Diane (2019). "Plants Are Not Animals and Animals Are Not Plants, Right? Wrong! Tiny Creatures in the Ocean Can be Both at Once!". Frontiers for Young Minds. 7. doi:10.3389/frym.2019.00048. S2CID 85528264.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Park, MG; Kim, S.; Kim, HS; Myung, G.; Kang, YG; Yih, W. (2006). "First successful culture of the marine dinoflagellate Dinophysis acuminata". Aquatic Microbial Ecology. 45: 101–106. doi:10.3354/ame045101.
- ^ Stoecker, D.; Tillmann, U.; Granéli, E. (2006). "Phagotrophy in Harmful Algae". Ecology of Harmful Algae. Ecological Studies. Vol. 189. pp. 177–187. doi:10.1007/978-3-540-32210-8_14. ISBN 978-3-540-32209-2.
- ^ a b Flynn, Kevin J.; Stoecker, Diane K.; Mitra, Aditee; Raven, John A.; Glibert, Patricia M.; Hansen, Per Juel; Granéli, Edna; Burkholder, Joann M. (2013). "Misuse of the phytoplankton–zooplankton dichotomy: The need to assign organisms as mixotrophs within plankton functional types". Journal of Plankton Research. 35: 3–11. doi:10.1093/plankt/fbs062.
- ^ a b Stoecker, Diane K. (1998). "Conceptual models of mixotrophy in planktonic protists and some ecological and evolutionary implications". European Journal of Protistology. 34 (3): 281–290. doi:10.1016/S0932-4739(98)80055-2.
- ^ Frost, B. W. (1972). "Effects of Size and Concentration of Food Particles on the Feeding Behavior of the Marine Planktonic Copepod Calanus Pacificus 1". Limnology and Oceanography. 17 (6): 805–815. Bibcode:1972LimOc..17..805F. doi:10.4319/lo.1972.17.6.0805.
- ^ Landry, M. R.; Hassett, R. P. (1982). "Estimating the grazing impact of marine micro-zooplankton". Marine Biology. 67 (3): 283–288. doi:10.1007/BF00397668. S2CID 54759937.
- ^ Sanders, Robert W.; Gast, Rebecca J. (2012). "Bacterivory by phototrophic picoplankton and nanoplankton in Arctic waters". FEMS Microbiology Ecology. 82 (2): 242–253. doi:10.1111/j.1574-6941.2011.01253.x. PMID 22092954.
- ^ Sherr, E. B., and B. F. Sherr (1993) "Protistan grazing rates via uptake of fluorescently labeled prey", pages 695–701. In: P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole [eds.], Handbook of methods in aquatic microbial ecology, Lewis.
- ^ Roman, Michael R.; Rublee, Parke A. (1980). "Containment effects in copepod grazing experiments: A plea to end the black box approach1". Limnology and Oceanography. 25 (6): 982–990. doi:10.4319/lo.1980.25.6.0982.
- ^ Steinkamp, J.; Wilson, J.; Saunders, G.; Stewart, C. (1982). "Phagocytosis: Flow cytometric quantitation with fluorescent microspheres". Science. 215 (4528): 64–66. Bibcode:1982Sci...215...64S. doi:10.1126/science.7053559. PMID 7053559.
- ^ Stickney, H.L.; Hood, R.R.; Stoecker, D.K. (2000). "The impact of mixotrophy on planktonic marine ecosystems". Ecological Modelling. 125 (2–3): 203–230. doi:10.1016/S0304-3800(99)00181-7.
- ^ Flynn, K. J.; Mitra, A. (2009). "Building the "perfect beast": Modelling mixotrophic plankton". Journal of Plankton Research. 31 (9): 965–992. doi:10.1093/plankt/fbp044.
- ^ Ward, Ben A.; Dutkiewicz, Stephanie; Barton, Andrew D.; Follows, Michael J. (2011). "Biophysical Aspects of Resource Acquisition and Competition in Algal Mixotrophs". The American Naturalist. 178 (1): 98–112. doi:10.1086/660284. hdl:1721.1/97884. PMID 21670581. S2CID 8396559.
- ^ d'Alelio, Domenico; Libralato, Simone; Wyatt, Timothy; Ribera d'Alcalà, Maurizio (2016). "Ecological-network models link diversity, structure and function in the plankton food-web". Scientific Reports. 6: 21806. Bibcode:2016NatSR...621806D. doi:10.1038/srep21806. PMC 4756299. PMID 26883643.
- ^ Seong, KA; Jeong, HJ; Kim, S.; Kim, GH; Kang, JH (2006). "Bacterivory by co-occurring red-tide algae, heterotrophic nanoflagellates, and ciliates". Marine Ecology Progress Series. 322: 85–97. Bibcode:2006MEPS..322...85S. doi:10.3354/meps322085.
- ^ Sanders, Robert W.; Porter, Karen G.; Bennett, Susan J.; Debiase, Adrienne E. (1989). "Seasonal patterns of bacterivory by flagellates, ciliates, rotifers, and cladocerans in a freshwater planktonic community". Limnology and Oceanography. 34 (4): 673–687. Bibcode:1989LimOc..34..673S. doi:10.4319/lo.1989.34.4.0673.
- ^ Unrein, Fernando; Massana, Ramon; Alonso-Sáez, Laura; Gasol, Josep M. (2007). "Significant year-round effect of small mixotrophic flagellates on bacterioplankton in an oligotrophic coastal system". Limnology and Oceanography. 52 (1): 456–469. Bibcode:2007LimOc..52..456U. doi:10.4319/lo.2007.52.1.0456. S2CID 59330965.
- ^ Domaizon, Isabelle; Viboud, Sylvie; Fontvieille, Dominique (2003). "Taxon-specific and seasonal variations in flagellates grazing on heterotrophic bacteria in the oligotrophic Lake Annecy – importance of mixotrophy". FEMS Microbiology Ecology. 46 (3): 317–329. doi:10.1016/S0168-6496(03)00248-4. PMID 19719562.
- ^ Verma, Arjun; Barua, Abanti; Ruvindy, Rendy; Savela, Henna; Ajani, Penelope A.; Murray, Shauna A. (2019). "The Genetic Basis of Toxin Biosynthesis in Dinoflagellates". Microorganisms. 7 (8): 222. doi:10.3390/microorganisms7080222. PMC 6722697. PMID 31362398..
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c d e f g h Flynn, Kevin J.; Mitra, Aditee; Glibert, Patricia M.; Burkholder, Joann M. (2018). "Mixotrophy in Harmful Algal Blooms: By Whom, on Whom, When, Why, and What Next". Global Ecology and Oceanography of Harmful Algal Blooms. Ecological Studies. Vol. 232. pp. 113–132. doi:10.1007/978-3-319-70069-4_7. ISBN 978-3-319-70068-7.
Myxobacteria
[edit]- Myxobacteria Are Able to Prey Broadly upon Clinically-Relevant Pathogens, Exhibiting a Prey Range Which Cannot Be Explained by Phylogeny
- Bacteriophages of Myxococcus xanthus, a Social Bacterium
Protists
[edit]

Box models
[edit]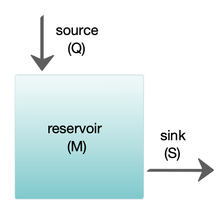
FROM: Climate model...
Box models are widely used to model biogeochemical systems.[3] Box models are simplified versions of complex systems, reducing them to boxes (or storage reservoirs) linked by fluxes. The boxes are assumed to behave as if they were mixed homogeneously. Simple box models have a small number of boxes with properties, such as volume, that do not change with time. These can often be used to derive analytical formulas describing the dynamics and steady-state abundance of the chemical species involved. More complex multibox models are usually solved using numerical techniques.
The diagram at the right shows a basic one-box model.
Box or reservoir models... the box is a storage reservoir... If the flow is steady (but a generalization to non-steady flow is possible[3]) and is conservative, then the exit age distribution and the internal age distribution can be related one to the other
τ0 = M/S where τ is the turnover time (renewal time, residence time or exit age), M is the mass in the reservoir, Q is the total flux into the reservoir, and S (= kM) is the total flux out of the reservoir. The equation describing the rate of change of content in a reservoir is dM/dt = Q - S = Q - M/0.
"Some of the first applications of the integrative field of "Biogeochemistry" are derived from organic geochemical studies where organisms and their molecular biochemistry were used as an initial framework for interpreting sources of sedimentary organic matter (Abelson and Hoering, 1960; Eglinton and Calvin, 1967. Biogeochemical cycles involve the interaction of biological, chemical, and geological processes that determine sources, sinks, and fluxes of elements through different reservoirs within ecosystems. Much of this book will use this basic box-model approach to understand the cycling of elements in estuarine systems." [2]
"Therefore, we need to first define some of the basic terms before we can understand how fluxes and reservoirs interact to determine chemical budgets in a biogeochemical box model (figure 1.4). For example, a reservoir is the amount of material (M), as defined by its chemical, physical, and/or biological properties. The units used to quantify material in a reservoir, in the box or compartment of a box model, are typically of mass or moles. Flux (F) is defined as the amount of material that is transported from one reservoir to another over a particular time period (mass/time or mass/area/time). A source Si is defined as the flux of material into a reservoir, while a sink So) is the flux of material out of the reservoir. The turnover time is required to remove all the materials in a reservoir, or the average time spent by elements in a reservoir."[2]
"Finally, a budget is essentially a "checks and balances" of all the sources and sinks as they relate to the material turnover in reservoirs. For example, if the sources and sinks are the same, and do not change over time, the reservoir is considered to he in a steady state".[2]
"The term cycle refers to when there are two or more connecting reservoirs, whereby materials are cycled through the system—generally with a predictable pattern of cyclic flow."[2]
"A one-box model for an atmospheric species X is shown in Figure 3-1 . It describes the abundance of X inside a box representing a selected atmospheric domain (which could be for example an urban area, the United States, or the global atmosphere). Transport is treated as a flow of X into the box (Fin) and out of the box (Fout). If the box is the global atmosphere then Fin = Fout = 0. The production and loss rates of X inside the box may include contributions from emissions (E), chemical production (P), chemical loss (L), and deposition (D). The terms Fin, E, and P are sources of X in the box; the terms Fout, L, and D are sinks of X in the box. The mass of X in the box is often called an inventory and the box itself is often called a reservoir. The one-box model does not resolve the spatial distribution of the concentration of X inside the box. It is frequently assumed that the box is well-mixed in order to facilitate computation of sources and sinks."[4]
"MULTI-BOX MODELS: The one-box model is a particularly simple tool for describing the chemical evolution of species in the atmosphere. However, the drastic simplification of transport is often unacceptable. Also, the model offers no information on concentration gradients within the box. A next step beyond the one-box model is to describe the atmospheric domain of interest as an assemblage of N boxes exchanging mass with each other."[4]
"The turnover time is the ratio between the content (M) of a reservoir and the total flux out of it (S). The residence time is the time spent in a reservoir by an individual atom or molecule. It is also the age of a molecule when it leaves the reservoir. If the pathway of the atom from the source to the sink is characterized by a physical transport, the term “transit time” can be used as an alternative. The age of an atom or molecule in a reservoir is the time since it entered the reservoir. Age is defined for all molecules, whether they are leaving the reservoir or not. The response time of a reservoir is a time-scale that characterizes the adjustment to equilibrium after a sudden change in the system. A linear system of reservoirs is one where the fluxes among the reservoirs are linearly related to the reservoir contents. A special case, that is commonly assumed to apply, is one where the fluxes among reservoirs are proportional to the content of the reservoirs where they originate."[5]
"The purpose of this chapter is to introduce the tools necessary to develop the two main types of models used in chemical oceanography. These are:. -Box (or reservoir) Models and -Continuous Transport-reaction Models ... - Reservoir (M)(also box or compartment) - The amount of material contained by a defined physical regime, such as the atmosphere, the surface ocean or the lithosphere. The size of the reservoirs are determined by the scale of the analysis as well as the homogeneity of the spatial distribution. The units are usually in mass of moles. - Flux (F) - The amount of material transferred from one reservoir to another per unit time. - Source (Q) - A flux of material into a reservoir. - Sink (S) - A flux of material out of a reservoir - Budget - A balance equation of all sources and sinks for a given reservoir. - Turnover Time (τ) - The ratio of the content (M) of a reservoir divided by the sum of its sources (ΣQ) or sinks (ΣS). Thus τ = M/ΣQ or τ = M/ΣS. - Cycle - A system consisting of two or more connected reservoirs where a large fraction of the material is transferred through the system in a cyclic fashion. Budgets and cycles can be considered over a wide range of spatial scales from local to global. - Steady State - When the sources and sinks are in balance and do not change with time. - Closed System - When all the material cycles within the system - Open System - When material exchanges with outside the system." PLUS LOTS MORE...[6]
Global biogeochemical box models usually measure:
— reservoir masses in petagrams (Pg)
— flow fluxes in petagrams per year (Pg yr−1)
These units are used in the diagrams below
____________________________________________
one petagram = 1015 grams = one gigatonne = one billion tonnes
In the box diagram below on the left, net phytoplankton production is about 50 Pg C (1 Pg = 1 Gt = 1015 g) each year, 10 Pg is exported to the ocean interior, the other 40 Pg is respired in the euphotic zone. Organic carbon degradation continues while particles settle through the ocean interior and only 2 Pg eventually arrives at the seafloor, the other 8 Pg is respired in the dark ocean. In sediments, the time scale available for degradation increases order of magnitude with the result that 90% of the organic carbon delivered is degraded and only 0.2 Pg C yr−1 is eventually buried and transferred from the biosphere to the geosphere.[7]


example of a more complex model with multiple interacting boxes
Sloppy feeding
[edit]2011: 65 citations... "Crustacean zooplankton produce dissolved organic matter (DOM) and inorganic nutrients via sloppy feeding, excretion, and fecal pellet leaching. These different mechanisms of the release of metabolic products, however, have never been individually isolated. Our study was designed to determine the relative importance of these different modes on release of dissolved organic carbon (DOC), ammonium (NH4+), and urea from Acartia tonsa calanoid copepods feeding on the diatom Thalassiosira weissflogii. Excretion and sloppy feeding were the dominant modes of DOC production (80 and 20% of total DOC release, respectively) and NH4+ release (93 and 7% of total NH4+ release, respectively). Urea, however, was predominately produced via sloppy feeding and fecal pellet leaching (25% and 62% of total urea release, respectively). Urea contributed 20% of total measured nitrogen (TMN; NH4+ + urea) released from copepods, and constituted 100% of TMN released via fecal pellet leaching, 47% of TMN released via sloppy feeding, and only 3.5% of TMN released via excretion. TMN release was > 100% of copepod body N d− 1, resulting in low DOC:TMN release ratios (4.1 for sloppy feeding, 2.1 for cumulative release of sloppy feeding, excretion, and fecal pellet leaching). Our results suggest that the mechanism of release plays an important role in the amount of different forms of DOM, NH4+, and urea available to bacteria and phytoplankton. Research highlights ► Relative importance of sloppy feeding, excretion, and fecal pellet leaching varied. ► Excretion and sloppy feeding were the dominant modes of DOC and NH4+ release. ► Sloppy feeding and fecal pellet leaching were dominant modes of urea release. ► Ingestion and sloppy feeding were highest in the first 20 min of the 3 h incubation. ► Excretion and fecal pellet leaching were consistently lower in non-feeding copepods." [11]
2017: 164 citations... "Marine zooplankton comprise a phylogenetically and functionally diverse assemblage of protistan and metazoan consumers that occupy multiple trophic levels in pelagic food webs. Within this complex network, carbon flows via alternative zooplankton pathways drive temporal and spatial variability in production-grazing coupling, nutrient cycling, export, and transfer efficiency to higher trophic levels. We explore current knowledge of the processing of zooplankton food ingestion by absorption, egestion, respiration, excretion, and growth (production) processes. On a global scale, carbon fluxes are reasonably constrained by the grazing impact of microzooplankton and the respiratory requirements of mesozooplankton but are sensitive to uncertainties in trophic structure. The relative importance, combined magnitude, and efficiency of export mechanisms (mucous feeding webs, fecal pellets, molts, carcasses, and vertical migrations) likewise reflect regional variability in community structure. Climate change is expected to broadly alter carbon cycling by zooplankton and to have direct impacts on key species." [12]
324 citations... Turner, J.T. (2015) "Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump". Progress in Oceanography, 130: 205–248. doi:10.1016/j.pocean.2014.08.005.</ref>
SLOPPY FEEDING in zooplankton Zooplankton and the Ocean Carbon Cycle "Zooplankton biogeochemical cycles"
LAMPERT "Lampert (1978) first demonstrated that as zooplankton graze on phytoplankton they damage the prey cell, releasing DOC via “sloppy feeding."[13]
SLOPPY FEEDING DEFINITION "Sloppy Feeding: Loss of prey biomass during feeding process. Note that sloppy feeding is a process associated with metazoans, not protists, since protists engulf their prey whole. Organisms that rip or tear their prey would contribute to sloppy feeding."
EGESTION "Egestion: Losses of non-digestable or partially digested material prior to assimilation • This material becomes part of the detritus pool in the euphotic zone (DOM), or • It is lost from euphotic zone as fecal transport (via fecal pellets) (POM)
EXUDATION
OTHER MECHANISMS "Sloppy feeding and exudation were long believed to be the major mechanisms, but with discoveries of new links in the food web there has also been recognition of additional mechanisms of DOM production. The list now includes microbial ectohydrolase activity, viral lysis of phytoplankton and heterotrophic microbes, cellular release of transparent exopolymer particles and other gel particles, programed cell death, and microbe-microbe antagonism".[14] . . . . . .
There has been significant speculation on other possible zooplankton- mediated mechanisms of DOM production from the ... Further, phytoplankton cell breakage and loss of cellular contents during macrozooplankton grazing (sloppy feeding) ...
However, as pointed out by Fogg (1969), elevated DOM may be a result (due to DOM excretion, bacterial and viral lysis, and zooplankton "sloppy feeding"), rather than a cause of cyanobacterial blooms.
In addition , a significant amount of the carbon consumed by crustacean zooplankton can be lost as DOC ( e . g . , Hargrave 1971 ; Dagg 1976 ) . DOC also can be released during " sloppy feeding " , excreted during ingestion , or can diffuse ...
Although neither viral attack (Wilhelm and Suttle, 1999) nor sloppy feeding (Lampert, 1978; Riemann et al., 1986; ..
Macrozooplankton contribute significantly to the POM by fecal pellet production , to the DIM by phosphate and ammonia excretion , and possibly , to the DOM through excretion and mechanisms such as sloppy feeding where phytoplankton ...
different or additional carbon sources were made available to bacteria by the grazing activities of zooplankton . An important mechanism providing additional carbon sources to the bacteria may be seen in “ sloppy feeding " ( LAMPERT 1978 ) ...
In the oceans and freshwaters, both herbivorous and carnivorous species comprise the zooplankton. ... Grazing may also be important in recycling nutrients released from cells broken during ingestion (“sloppy feeding”) or excreted in wastes.
For example, extensive studies on excretion and defecation by zooplankton have been conducted (Conover, 1966; ... dissolved forms associated with so-called sloppy feeding and other feeding mechanisms would occur simultaneously, these ...
Møller (2007) found sloppy feeding to be a function of the copepod-prey size ratio when prey size was relatively large (ratio ... Food-chain effects may also be observed, such as selective predation upon protozooplankton by copepods in the ...
Deposition of particulate organic matter, such as phytoplankton, within the sediment can be substantial in the presence of suspension-feeders, either by sloppy feeding of the captured particles or secondarily by organic-rich mucus secretions ...
This material is exuded from living phytoplankton cells, released from the front end (sloppy feeding) and TABLE 10.4 Comparisons of percent primary production grazed per day by protists based on a synthesis of results from paire
Algal cell breakage during herbivore grazing ('sloppy feeding') leads to DOM release which can not be distinguished from exudation. Respiration of ingested carbon by herbivores may lead to underestimation of exudation. Although the extent ...
DEBORAH STEINBERG "Crustacean zooplankton produce dissolved organic matter (DOM) and inorganic nutrients via sloppy feeding, excretion, and fecal pellet leaching. These different mechanisms of the release of metabolic products, however, have never been individually isolated."[11]
- Steinberg, D. (2017) https://books.google.co.nz/books?id=NuUWDgAAQBAJ&pg=PA52&dq=%22sloppy+feeding%22+zooplankton&hl=en&newbks=1&newbks_redir=0&sa=X&ved=2ahUKEwjzivf665rqAhVHdCsKHQHxDz0QuwUwAHoECAEQBg#v=onepage&q=%22sloppy%20feeding%22%20zooplankton&f=false "Zooplankton biogeochemical cycles". In: Castellani C and Edwards M (Eds.) Marine Plankton: A Practical Guide to Ecology, Methodology, and Taxonomy, pages 52–66, Oxford University Press. ISBN 9780199233267.
- Steinberg, D.K. and Landry, M.R. (2017) "Zooplankton and the ocean carbon cycle". Annual Review of Marine Science, 9: 413-444. doi:10.1146/annurev-marine-010814-015924.
"Marine zooplankton comprise a phylogenetically and functionally diverse assemblage of protistan and metazoan consumers that occupy multiple trophic levels in pelagic food webs. Within this complex network, carbon flows via alternative zooplankton pathways drive temporal and spatial variability in production-grazing coupling, nutrient cycling, export, and transfer efficiency to higher trophic levels. We explore current knowledge of the processing of zooplankton food ingestion by absorption, egestion, respiration, excretion, and growth (production) processes. On a global scale, carbon fluxes are reasonably constrained by the grazing impact of microzooplankton and the respiratory requirements of mesozooplankton but are sensitive to uncertainties in trophic structure. The relative importance, combined magnitude, and efficiency of export mechanisms (mucous feeding webs, fecal pellets, molts, carcasses, and vertical migrations) likewise reflect regional variability in community structure. Climate change is expected to broadly alter carbon cycling by zooplankton and to have direct impacts on key species."[15]
- ^ a b Colin, S., Coelho, L.P., Sunagawa, S., Bowler, C., Karsenti, E., Bork, P., Pepperkok, R. and De Vargas, C. (2017) "Quantitative 3D-imaging for cell biology and ecology of environmental microbial eukaryotes". eLife, 6: e26066. doi:10.7554/eLife.26066.002.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c d e Bianchi, Thomas (2007) Biogeochemistry of Estuaries page 9, Oxford University Press. ISBN 9780195160826.
- ^ Sarmiento, J.L.; Toggweiler, J.R. (1984). "A new model for the role of the oceans in determining atmospheric P CO 2". Nature. 308 (5960): 621–24. Bibcode:1984Natur.308..621S. doi:10.1038/308621a0. S2CID 4312683.
- ^ a b Jacob, Daniel J. (1999) Introduction to Atmospheric Chemistry, Chapter 3: Simple models, pages 24–40, Princeton University Press. ISBN 9780691001852. pdf version
- ^ Rodhe, H. (1992) "4 Modeling Biogeochemical Cycles". In: International Geophysics, Volume 50, pages 55–72, Academic Press.
- ^ Murray, James W. (2004) "Mass Balance - The Cornerstone of Chemical Oceanography". University of Washington.
- ^ a b Middelburg, J.J.(2019) Marine carbon biogeochemistry: a primer for earth system scientists, page 5, Springer Nature. ISBN 9783030108229. doi:10.1007/978-3-030-10822-9.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Sarmiento, Jorge L.; Gruber, Nicolas (2002). "Sinks for Anthropogenic Carbon". Physics Today. 55 (8): 30–36. Bibcode:2002PhT....55h..30S. doi:10.1063/1.1510279. S2CID 128553441.
- ^ Chhabra, Abha (2013). "Carbon and Other Biogeochemical Cycles". doi:10.13140/2.1.1081.8883.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Kandasamy, Selvaraj; Nagender Nath, Bejugam (2016). "Perspectives on the Terrestrial Organic Matter Transport and Burial along the Land-Deep Sea Continuum: Caveats in Our Understanding of Biogeochemical Processes and Future Needs". Frontiers in Marine Science. 3. doi:10.3389/fmars.2016.00259. S2CID 30408500.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Saba, G.K., Steinberg, D.K. and Bronk, D.A. (2011) "The relative importance of sloppy feeding, excretion, and fecal pellet leaching in the release of dissolved carbon and nitrogen by Acartia tonsa copepods". Journal of Experimental Marine Biology and Ecology, 404(1–2): 47–56. doi:10.1016/j.jembe.2011.04.013.
- ^ Steinberg, D.K. and Landry, M.R. (2017) "Zooplankton and the ocean carbon cycle". Annual Review of Marine Science, 9: 413–444. doi:10.1146/annurev-marine-010814-01592410.1146/annurev-marine-010814-015924.
- ^ Lampert W. (1978) "Release of dissolved organic carbon by grazing zooplankton". Limnology and Oceanography, 23(4): 831-834. doi:10.4319/lo.1978.23.4.0831.
- ^ Hansell DA and Carlson CA (2014) Biogeochemistry of Marine Dissolved Organic Matter Academic Press, pages 6–7. ISBN 9780124071537.
- ^ Steinberg, D.K. and Landry, M.R. (2017) "Zooplankton and the ocean carbon cycle". Annual Review of Marine Science, 9: 413-444. doi:10.1146/annurev-marine-010814-015924.
Viral lysis
[edit]"Microorganisms dominate the oceans and exert considerable control over fluxes of nutrients, organic mat- ter, and energy. This control is intimately related to the life cycles of these organisms, and whereas we know much about their modes and rates of reproduction, comparably little is known about how they die. The method of death for a microorganism is a primary factor controlling the fate of the nutrients and organic matter in the cell, e.g., whether they are incorporated into higher trophic levels, sink out of the water column, or are recy- cled within the microbial loop. This review addresses the different sources of mortality for marine microbes including grazing, viral lysis, programmed cell death, and necrosis. We describe each mode of death, the methods used to quantify them, what is known of their relative importance in the ocean, and how these vectors of mortality differentially affect the flow of organic matter in the open ocean. We then conclude with an assessment of how these forms of mortality are incorporated into current numerical ecosystem models and suggest future avenues of research to increase our understanding of the effects of death processes in oceanic food webs."[1]
Algae and cyanobacteria
[edit]"Plants means land plants. Algae means all eukaryotes with plastids, except plants. Some algae are protists. Some algae have lost the ability to photosynthesize. Algae are polyphyletic. Plants are monophyletic, and are descended from an alga. Plastids are monophyletic, and are descended from a cyanobacterium through endosymbiosis. The monophyly of plastids requires secondary endosymbiosis, at least... Red, green, and blue algae have primary plastids, descended from a cyanobacterium through the unique primary endosymbiotic event. All other algae have secondary or tertiary plastids, derived from red or green algae through secondary or tertiary endosymbiosis. Plants have primary plastids because plants are descended from a green alga. The group with primary plastids is monophyletic. Paulinella is a freak.
Life on Earth depends on cyanobacteria. Also, it was the free oxygen produced by cyanobacteria that made eukaryotes possible in the first place. Cyanobacteria did change the world. Twice." – user comments
Marine biogeochemical cycles
[edit]"The ocean is a great store of chemicals that receives inputs from rivers and the atmosphere and, on average, loses equal amounts to sedimentary deposits on the ocean floor." - Encyclopædia Britannica
Background
[edit]"The biosphere is made up of three main elements - hydrogen (49.8 per cent by weight), oxygen (24.9 per cent) and carbon (24.9 per cent). Several other elements are found in the biosphere, and some of them are essential to biological processes – nitrogen (0.27 per cent), calcium (0.073 per cent), potassium (0.046 per cent)silicon (0.033per cent), magnesium (0.031 per cent), phosphorus (0.030 per cent), sulphur (0.017 per cent), and aluminium (0.016 per cent). These elements, except aluminium, are the basic ingredients for organic compounds around which biochemistry revolves. Carbon, hydrogen, nitrogen, oxygen, sulphur and phosphorus are needed to build nucleic acids (RNA and DNA). amino acids (proteins), carbohydrates (sugars, starches and cellulose) and lipids (fats and fat-like materials). Calcium, magnesium and potassium are required in moderate amounts."[2]
| Element | Percent by mass |
|---|---|
| Oxygen | 85.84 |
| Hydrogen | 10.82 |
| Chlorine | 1.94 |
| Sodium | 1.08 |
| Magnesium | 0.1292 |
| Sulfur | 0.091 |
| Calcium | 0.04 |
| Potassium | 0.04 |
| Bromine | 0.0067 |
| Carbon | 0.0028 |
| Vanadium | 1.5 × 10−11 – 3.3 × 10−11 |
| Gas | Volume(A) | ||
|---|---|---|---|
| Name | Formula | in ppmv(B) | in % |
| Nitrogen | N2 | 780,840 | 78.084 |
| Oxygen | O2 | 209,460 | 20.946 |
| Argon | Ar | 9,340 | 0.9340 |
| Carbon dioxide (April, 2020)(C)[4] |
CO2 | 413.61 | 0.041361 |
| Neon | Ne | 18.18 | 0.001818 |
| Helium | He | 5.24 | 0.000524 |
| Methane | CH4 | 1.87 | 0.000187 |
| Krypton | Kr | 1.14 | 0.000114 |
| Not included in above dry atmosphere: | |||
| Water vapor(D) | H2O | 0–30,000(D) | 0–3%(E) |
| notes: (A) volume fraction is equal to mole fraction for ideal gas only, | |||
"Chemical elements required in moderate and large quantities are macronutrients. More than a dozen elements are required in trace amounts, including chlorine, chromium, copper, cobalt, iodine iron, manganese, molybdenum, nickel, selenium, sodium, vanadium and zinc. These are micronutrients... Other mineral elements cycle through living systems but have no known metabolic role."[2]
There is in the ecosphere a constant turnover of chemicals. The motive force behind these chemical cycles is life. In addition, on geological timescales, forces in the geosphere producing and consuming rocks influence the cycles. Biogeochemical cycles, as they are called, involve the storage and flux of all terrestrial elements and compounds except the inert ones. Material exchanges between life and life support systems are part of the biogeochemical cycles."[2]
"At their grandest scale, biogeochemical cycles involve the entire Earth. An exogenic cycle, involving the transport and transformation of materials near the Earth's surface, is normally distinguished from a slower and less well understood endogenic cycle involving the lower crust and mantle. Cycles of carbon, hydrogen, oxygen and nitrogen are gaseous cycles – their component chemical species are gaseous for a leg of the cycle. Other chemical species follow sedimentary cyclesbecause they do not readily volatilise and are exchanged between the biosphere and its environment in solution."[2]
- substance turnover - ecosystem turnover - cycling of substances - recycling machines
"A biogeochemical cycle defines the pathways by which chemical elements occurring in organisms are cycled between different living and nonliving compartments on the Earth (e.g., biosphere, atmosphere, hydrosphere, geosphere, etc.). Typical driving forces of a biogeochemical cycle are the metabolisms of living organisms, geological processes, or chemical reactions".[6]
"Water contains hydrogen and oxygen, which is essential to all living processes. The hydrosphere is the area of the Earth where water movement and storage occurs: as liquid water on the surface and beneath the surface or frozen (rivers, lakes, oceans, groundwater, polar ice caps, and glaciers), and as water vapor in the atmosphere. Carbon is found in all organic macromolecules and is an important constituent of fossil fuels. Nitrogen is a major component of our nucleic acids and proteins and is critical to human agriculture. Phosphorus, a major component of nucleic acid (along with nitrogen), is one of the main ingredients in artificial fertilizers used in agriculture and their associated environmental impacts on our surface water. Sulfur, critical to the 3–D folding of proteins (as in disulfide binding), is released into the atmosphere by the burning of fossil fuels, such as coal."[7]
"The cycling of these elements is interconnected. For example, the movement of water is critical for the leaching of nitrogen and phosphate into rivers, lakes, and oceans. Furthermore, the ocean itself is a major reservoir for carbon. Thus, mineral nutrients are cycled, either rapidly or slowly, through the entire biosphere, from one living organism to another, and between the biotic and abiotic world."[7]
- function of microorganisms
bacteria move minerals from inorganic states to organic states
"Here, we will focus on the function of microorganisms in these cycles, which play roles at each step. The reactions in these cycles are all redox reactions: in one direction, a compound is oxidized, and in the reverse direction, it is reduced. Because some of those reactions are primarily, or exclusively, performed by the microbes, specifically the bacteria and archaea, the microbes are critical to the continued function of the biosphere[8] and have been described as the "biological infrastructure" of the planet."[9]
"Oxygenic photosynthetic progenitors transformed the world's atmosphere from anoxic to oxic during the Great Oxygenation Event, beginning approximately 3,000,000,000 years ago [11]. Prochlorococcus and Synechococcus are now two of the world's most abundant cellular life forms, filling the ocean to varying degrees from pole to pole, generating oxygen as a byproduct of sunlight-driven photosynthesis. If these great oxygen sources vanished from the world's oceans, lakes, surface soils, and plant surfaces, then what would happen? Perhaps surprisingly, it is unlikely that anything problematic for aerobic life would happen for at least a few hundred thousand years. Assuming humans could distribute nitrogen globally, algae and plants could be expected to continue generating a proportion of available atmospheric oxygen, potentially as high as 50% [12]. Existing pools of atmospheric oxygen might satisfy the demand for aerobic metabolism among surviving organisms, possibly for decades or centuries. If this were the case, then asphyxiation of aerobic life would not be likely in the near term."[8]
"Microbes drive the nutrient and energy transformations that sustain Earth’s ecosystems,[10] and the viruses that infect them modulate both microbial population size and diversity".[11][12]
According to Farooq Azam and Francesca Malfatti, "Microbial oceanography is a field that is caught between scales — microbial processes must be understood at the scale of the individual microorganism, but yet we want to understand the cumulative influence of microbial processes on how the ocean works as a biogeochemical system... understanding how bacteria interact with the ocean system at the nanometre to millimetre scales provides insights into biogeochemical processes of global significance."[13]
- Mineral ecology
FROM: mineral (nutrient)
Minerals can be bioengineered by bacteria which act on metals to catalyze mineral dissolution and precipitation.[14] Mineral nutrients are recycled by bacteria distributed throughout soils, oceans, freshwater, groundwater, and glacier meltwater systems worldwide.[14][15] Bacteria absorb dissolved organic matter containing minerals as they scavenge phytoplankton blooms.[15] Mineral nutrients cycle through this marine food chain, from bacteria and phytoplankton to flagellates and zooplankton, which are then eaten by other marine life.[14][15] In terrestrial ecosystems, fungi have similar roles as bacteria, mobilizing minerals from matter inaccessible by other organisms, then transporting the acquired nutrients to local ecosystems.[16][17]
The biological carbon pump begins with phytoplankton and ends with sinking poo, eg, barnacle nauplius.

Nutrient cycles
[edit]FROM: nutrient cycle, eutrophication
- Transport of nutrients and minerals
Land interactions impact marine food webs in many ways. Coastlines typically have continental shelves extending some way from the shore. These provide extensive shallows sunlit down to the seafloor, allowing for photosynthesis and enabling habitats for seagrass meadows, coral reefs, kelp forests and other benthic life. Further from shore the continental shelf slopes towards deep water. Wind blowing at the ocean surface or deep ocean currents can result in cold and nutrient rich waters from abyssal depths moving up the continental slopes. This can result in upwellings along the outer edges of continental shelves, providing conditions for phytoplankton blooms.
Water evaporated by the sun from the surface of the ocean can precipitate on land and eventually return to the ocean as runoff or discharge from rivers, enriched with nutrients as well as pollutants. As rivers discharge into estuaries, freshwater mixes with saltwater and becomes brackish. This provides another shallow water habitat where mangrove forests and estuarine fish thrive. Overall, life in inland lakes can evolve with greater diversity than happens in the sea, because freshwater habitats are themselves diverse and compartmentalised in a way marine habitats are not. Some aquatic life, such as salmon and eels, migrate back and forth between freshwater and marine habitats. These migrations can result in the spread of pathogens and have impacts on the way life evolves in the ocean.
The ocean "transports heat from the equator to the poles, regulating the climate and weather patterns".[19]

"The subpolar Southern Ocean, south of the Antarctic Circumpolar Current (ACC), is arguably the most important region on Earth for the cycling of carbon on centennial to millennial time scales (1). Atmospheric carbon dioxide (CO2) exhibits strong sensitivity to the physical and biogeochemical dynamics of the region in paleoclimatic observations (2), low-complexity box models (3, 4), and fully three-dimensional numerical simulations (5, 6). Furthermore, in the present day, the regional uptake of anthropogenic carbon is a major contributor to its storage in the deep ocean (7, 8). Consequently, understanding the dynamics and variability of the subpolar Southern Ocean carbon cycle, and the system’s susceptibility to change, is a fundamental challenge facing modern climate research... Global climate is critically sensitive to physical and biogeochemical dynamics in the subpolar Southern Ocean, since it is here that deep, carbon-rich layers of the world ocean outcrop and exchange carbon with the atmosphere. Here, we present evidence that the conventional framework for the subpolar Southern Ocean carbon cycle, which attributes a dominant role to the vertical overturning circulation and shelf-sea processes, fundamentally misrepresents the drivers of regional carbon uptake. Observations in the Weddell Gyre—a key representative region of the subpolar Southern Ocean—show that the rate of carbon uptake is set by an interplay between the Gyre’s horizontal circulation and the remineralization at mid-depths of organic carbon sourced from biological production in the central gyre. These results demonstrate that reframing the carbon cycle of the subpolar Southern Ocean is an essential step to better define its role in past and future climate change."[21]
- Eutrophication
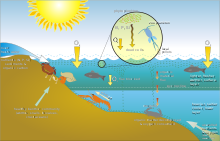
Eutrophication is a common phenomenon in coastal waters. In contrast to freshwater systems where phosphorus is often the limiting nutrient, nitrogen is more commonly the key limiting nutrient of marine waters; thus, nitrogen levels have greater importance to understanding eutrophication problems in salt water.[22] Estuaries, as the interface between freshwater and saltwater, can be both phosphorus and nitrogen limited and commonly exhibit symptoms of eutrophication. Eutrophication in estuaries often results in bottom water hypoxia/anoxia, leading to fish kills and habitat degradation.[22] Upwelling in coastal systems also promotes increased productivity by conveying deep, nutrient-rich waters to the surface, where the nutrients can be assimilated by algae. Examples of anthropogenic sources of nitrogen-rich pollution to coastal waters include seacage fish farming and discharges of ammonia from the production of coke from coal.
The World Resources Institute has identified 375 hypoxic coastal zones in the world, concentrated in coastal areas in Western Europe, the Eastern and Southern coasts of the US, and East Asia, particularly Japan.[23]
In addition to runoff from land, fish farming wastes and industrial ammonia discharges, atmospheric fixed nitrogen can be an important nutrient source in the open ocean. A study in 2008 found that this could account for around one third of the ocean's external (non-recycled) nitrogen supply, and up to 3% of the annual new marine biological production.[24] It has been suggested that accumulating reactive nitrogen in the environment may prove as serious as putting carbon dioxide in the atmosphere.[25]
When an ecosystem experiences an increase in nutrients, primary producers reap the benefits first. In aquatic ecosystems, species such as algae experience a population increase (called an algal bloom). Algal blooms limit the sunlight available to bottom-dwelling organisms and cause wide swings in the amount of dissolved oxygen in the water. Oxygen is required by all aerobically respiring plants and animals and it is replenished in daylight by photosynthesizing plants and algae. Under eutrophic conditions, dissolved oxygen greatly increases during the day, but is greatly reduced after dark by the respiring algae and by microorganisms that feed on the increasing mass of dead algae. When dissolved oxygen levels decline to hypoxic levels, fish and other marine animals suffocate. As a result, creatures such as fish, shrimp, and especially immobile bottom dwellers die off.[26] In extreme cases, anaerobic conditions ensue, promoting growth of bacteria. Zones where this occurs are known as dead zones.
- Ocean currents, conveyor belt etc

- effects of salinity on the ocean conveyor belt
"That’s a fairly complex question. The idea is that the conveyor belt has warm waters coming up in the Gulf Stream and entering the North Atlantic, losing their heat to the atmosphere, getting cold and dense and sinking, and then returning underneath to the tropics in the Atlantic. Now the idea is that the glacial melt could freshen that water and make it less dense and less likely to sink. But we’re looking at another part of the system that feeds the warm water sources that are heading northward. We’re finding that those are getting saltier. It may work out that the higher salinities that we are seeing in the subtropical underwater that Eric mentioned could counteract the freshening due to more precipitation and glacial melt at high latitudes. So it is a delicate balance. What we think now is that it’s not to likely that the conveyor belt is going to shut down any time soon. It might slow down a little bit from general warming, but we’re not going to see a shutdown such as happened in the past." - Raymond Schmitt 2013 [5]
-
Upwelling
- "Nutrient cycles... give our bodies energy and help our cells function... Nutrients move between living things, into the Earth, and into the atmosphere. This process is called a nutrient cycle. Things we need to survive like carbon-containing compounds such as sugar, micronutrients like nitrogen, phosphorus, and sulfur, and water, move through living things and our environment. Today we're going to look at the nutrient cycle for each of these important compounds."[6]
- "A nutrient cycle refers to the movement and exchange of organic and inorganic matter back into the production of living matter. The process is regulated by the food web pathways previously presented, which decompose organic matter into inorganic nutrients. Nutrient cycles occur within ecosystems. Nutrient cycles that we will examine in this section include water, carbon, oxygen and nitrogen cycles."[7]
Sulphur cycle
[edit]
Beggiatoa species inhabit marine mangrove sediments [28]
Iron cycle and dust
[edit]Calcium and silica cycles
[edit]Biomineralization
[edit]Deep water cycle
[edit]Fossil fuels
[edit]Other cycles
[edit]PCB cycle
[edit]Mercury cycle
[edit]
Rock cycle
[edit]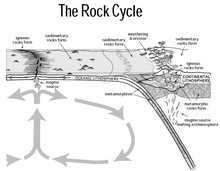
References
[edit]- ^ Brum JR, Morris JJ, Décima M and Stukel MR (2014) "Mortality in the oceans: Causes and consequences". Eco-DAS IX Symposium Proceedings, Chapter 2, pages 16-48. Association for the Sciences of Limnology and Oceanography. doi:10.4319/ecodas.2014.978-0-9845591-3-8.16. ISBN 978-0-9845591-3-8.
- ^ a b c d Huggett RJ (2004) Fundamentals of Biogeography, second edition, page 228, Routledge. ISBN 9781134349685.
- ^ Cite error: The named reference
total%was invoked but never defined (see the help page). - ^ "Vital signs: Carbon Dioxide". NASA Climate. May 2020. Retrieved 5 June 2020.
{{cite web}}: CS1 maint: url-status (link) - ^ Cite error: The named reference
WallaceHobbswas invoked but never defined (see the help page). - ^ Reitner, J. and Thiel, V. (Eds) (2011) Encyclopedia of Geobiology, page 137, Amsterdam: Springer. ISBN 9781402092114.
- ^ a b Cite error: The named reference
OpenStaxwas invoked but never defined (see the help page). - ^ a b Gilbert, J.A. and Neufeld, J.D. (2014) "Life in a world without microbes". PLoS Biology, 12(12): e1002020. doi:10.1371/journal.pbio.1002020.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Biogeochemical Cycles OpenStax, Microbiology.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Falkowski, P.G., Fenchel, T. and Delong, E.F.(2008) "The microbial engines that drive Earth's biogeochemical cycles". Science, 320(5879): 1034–1039. doi:10.1126/science.1153213
- ^ Brum, J.R. and Sullivan, M.B.(2015) "Rising to the challenge: accelerated pace of discovery transforms marine virology". Nature Reviews Microbiology, 13(3): 147–159. doi:10.1038/nrmicro3404
- ^ Murata, K., Zhang, Q., Galaz-Montoya, J.G., Fu, C., Coleman, M.L., Osburne, M.S., Schmid, M.F., Sullivan, M.B., Chisholm, S.W. and Chiu, W. (2017) "Visualizing adsorption of cyanophage P-SSP7 onto marine Prochlorococcus". Scientific Reports, 7: 44176. doi:10.1038/srep44176
- ^ Azam, F. and Malfatti, F. (2007) "Microbial structuring of marine ecosystems". Nature Reviews Microbiology, 5(10): 782–791. doi:10.1038/nrmicro1747.
- ^ a b c Warren LA, Kauffman ME (February 2003). "Geoscience. Microbial geoengineers". Science. 299 (5609): 1027–9. doi:10.1126/science.1072076. PMID 12586932. S2CID 19993145.
- ^ a b c Azam, F; Fenchel, T; Field, JG; Gray, JS; Meyer-Reil, LA; Thingstad, F (1983). "The ecological role of water-column microbes in the sea" (PDF). Mar. Ecol. Prog. Ser. 10: 257–63. Bibcode:1983MEPS...10..257A. doi:10.3354/meps010257.
- ^ J. Dighton (2007). "Nutrient Cycling by Saprotrophic Fungi in Terrestrial Habitats". In Kubicek, Christian P.; Druzhinina, Irina S (eds.). Environmental and microbial relationships (2nd ed.). Berlin: Springer. pp. 287–300. ISBN 978-3-540-71840-6.
- ^ Gadd GM (January 2017). "The Geomycology of Elemental Cycling and Transformations in the Environment" (PDF). Microbiol Spectr. 5 (1). doi:10.1128/microbiolspec.FUNK-0010-2016. PMID 28128071.
- ^ Shen, Y. and Benner, R. (2018) "Mixing it up in the ocean carbon cycle and the removal of refractory dissolved organic carbon". Scientific Reports, 8(1): 1–9. doi:10.1038/s41598-018-20857-5
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Why should we care about the ocean? NOAA: National Ocean Service. Updated: 7 January 2020. Retrieved 1 March 2020.
- ^ Bear R and Rintoul D (2018) "Biogeochemical Cycles". In: Bear R, Rintoul D, Snyder B, Smith-Caldas M, Herren C and Horne E (Eds) Principles of Biology OpenStax.
- ^ MacGilchrist, G.A., Garabato, A.C.N., Brown, P.J., Jullion, L., Bacon, S., Bakker, D.C., Hoppema, M., Meredith, M.P. and Torres-Valdés, S. (2019) "Reframing the carbon cycle of the subpolar Southern Ocean". Science advances, 5(8): eaav6410. doi:10.1126/sciadv.aav6410.
- ^ a b Paerl, Hans W.; Valdes, Lexia M.; Joyner, Alan R.; Piehler, Michael F.; Lebo, Martin E. (2004). "Solving problems resulting from solutions: Evolution of a dual nutrient management strategy for the eutrophying Neuse River Estuary, North Carolina". Environmental Science and Technology. 38 (11): 3068–3073. doi:10.1021/es0352350. PMID 15224737.
- ^ Selman, Mindy (2007) Eutrophication: An Overview of Status, Trends, Policies, and Strategies. World Resources Institute.
- ^ Duce, R A; et al. (2008). "Impacts of Atmospheric Anthropogenic Nitrogen on the Open Ocean". Science. 320 (5878): 893–89. Bibcode:2008Sci...320..893D. doi:10.1126/science.1150369. hdl:21.11116/0000-0001-CD7A-0. PMID 18487184. S2CID 11204131.
- ^ Addressing the nitrogen cascade Eureka Alert, 2008.
- ^ Horrigan, L.; Lawrence, R. S.; Walker, P. (2002). "How sustainable agriculture can address the environmental and human health harms of industrial agriculture". Environmental Health Perspectives. 110 (5): 445–456. doi:10.1289/ehp.02110445. PMC 1240832. PMID 12003747.
- ^ Tsai Y, Sawaya MR, Cannon GC; et al. (2007). "Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome". PLOS Biol. 5 (6): e144. doi:10.1371/journal.pbio.0050144. PMC 1872035. PMID 17518518.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Jean, M.R., Gonzalez-Rizzo, S., Gauffre-Autelin, P., Lengger, S.K., Schouten, S. and Gros, O. (2015) "Two new Beggiatoa species inhabiting marine mangrove sediments in the Caribbean". PLoS One, 10(2): e0117832. doi:10.1371/journal.pone.0117832.s001.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.

-
Bathymetry of the ocean floor showing the continental shelves (salmon colour) and the mid-ocean ridges (yellow-green)
-
Salinity [2]
Ocean carbon cycle
[edit]
- Carbonate–silicate cycle
- Biogeochemical cycles - wikibooks
Plankton is the engine for the biological pump, a process in which carbon is transported through the bodies of the organisms into the deep sea. This processes are important in view of the carbon sink discussion and hope for the future that some of the carbon currently accumulating in the Earth's atmosphere as carbon dioxide might be stored in the ocean.
-
Marine carbon cycle
-
Marine export production
-
Roles of fungi in the marine carbon cycle
-
Carbon cycle
-
Carbon cycle
-
Carbon cycle
-
Ocean carbon cycle
-
Carbon cycle flux
-
Carbon sequestration
-
Climate change carbon cycle
-
Reservoirs and fluxes of carbon in deep Earth
-
Carbon cycle-simple diagram
-
Subduction - accretionary
-
Subduction - Trench Schematic
-
Carbon dioxide emissions global carbon cycle



"The cycling of carbon on Earth exerts a fundamental influence upon the greenhouse gas content of the atmosphere, and hence global climate over millennia. Until recently, ice sheets were viewed as inert components of this cycle and largely disregarded in global models. Research in the past decade has transformed this view, demonstrating the existence of uniquely adapted microbial communities, high rates of biogeochemical/physical weathering in ice sheets and storage and cycling of organic carbon (>104 Pg C) and nutrients."[4]

Silica cycle
[edit]
- From
- Silica cycle...
Siliceous organisms in the ocean, such as diatoms and radiolaria, are the primary sink of dissolved silicic acid into opal silica.[12] Once in the ocean, dissolved Si molecules are biologically recycled roughly 25 times before export and permanent deposition in marine sediments on the seafloor.[15] This rapid recycling is dependent on the dissolution of silica in organic matter in the water column, followed by biological uptake in the photic zone. The estimated residence time of the silica biological reservoir is about 400 years.[15] Opal silica is predominately undersaturated in the world's oceans. This undersaturation promotes rapid dissolution as a result of constant recycling and long residence times. The estimated turnover time of Si is 1.5x104 years.[11] The total net inputs and outputs of silica in the ocean are 9.4 ± 4.7 Tmol Si yr−1 and 9.9 ± 7.3 Tmol Si yr−1, respectively.[11]
Biogenic silica production in the photic zone is estimated to be 240 ± 40 Tmol Si year −1.[11] Dissolution in the surface removes roughly 135 Tmol Si year−1, while the remaining Si is exported to the deep ocean within sinking particles.[15] In the deep ocean, another 26.2 Tmol Si Year−1 is dissolved before being deposited to the sediments as opal rain.[15] Over 90% of the silica here is dissolved, recycled and eventually upwelled for use again in the euphotic zone.[15]
-
Silica cycle
-
Forms of carbon in deep Earth
-
Carbon-Slicate Cycle Feedbacks
Oxygen cycle
[edit]
-
Oxygen cycle
-
Oxygen cycle
- Limburg, K.E., Breitburg, D., Swaney, D.P. and Jacinto, G. (2020) "Ocean Deoxygenation: A Primer. One Earth", 2(1): 24–29. doi:10.1038/s41598-018-27793-4
Ocean ventilation and deoxygenation
[edit]
"Changes of ocean ventilation rates and deoxygenation are two of the less obvious but important indirect impacts expected as a result of climate change on the oceans. They are expected to occur because of (i) the effects of increased stratification on ocean circulation and hence its ventilation, due to reduced upwelling, deep-water formation and turbulent mixing, (ii) reduced oxygenation through decreased oxygen solubility at higher surface temperature, and (iii) the effects of warming on biological production, respiration and remineralization. The potential socio-economic consequences of reduced oxygen levels on fisheries and ecosystems may be far-reaching and significant... Our current understanding of both the causes and consequences of reduced oxygen in the ocean, and our ability to represent them in models are therefore inadequate, and the reasons for this remain unclear. It is too early to say whether or not the socio-economic consequences are likely to be serious. However, the consequences are ecologically, biogeochemically and climatically potentially very significant, and further research on these indirect impacts of climate change via reduced ventilation and oxygenation of the oceans should be accorded a high priority."[17]
- Oxygen minimum zone
- Microbiology of oxygen minimum zones
- Watson, A.J., Lenton, T.M. and Mills, B.J. (2017) "Ocean deoxygenation, the global phosphorus cycle and the possibility of human-caused large-scale ocean anoxia". Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 375: 20160318. doi:10.1098/rsta.2016.0318.


Nutrient cycles
[edit]




Nitrogen cycle
[edit]"Nitrogen (N) is a key element for life in the oceans. It controls primary productivity in many parts of the global ocean, consequently playing a crucial role in the uptake of atmospheric carbon dioxide. The marine N cycle is driven by multiple biogeochemical transformations mediated by microorganisms, including processes contributing to the marine fixed N pool (N2 fixation) and retained N pool (nitrification, assimilation, and dissimilatory nitrate reduction to ammonia), as well as processes contributing to the fixed N loss (denitrification, anaerobic ammonium oxidation and nitrite-dependent anaerobic methane oxidation). The N cycle maintains the functioning of marine ecosystems and will be a crucial component in how the ocean responds to global environmental change. In this review, we summarize the current understanding of the marine microbial N cycle, the ecology and distribution of the main functional players involved, and the main impacts of anthropogenic activities on the marine N cycle."[24]



-
Marine nitrogen cycle
Phosphorus cycle
[edit]Sulfur cycle
[edit]-
Sulfur cycle
-
Sulfur cycle
-
Sulfur cycle
-
Sulfur cycle
Water cycle
[edit]-
Water cycle [3]
-
hydrological cycle
Rock cycle
[edit]-
Rock cycle in Wilson cycle
-
Rock cycle
-
Rock cycle
-
Rock cycle
Mercury cycle
[edit]Biogeochemical proxies
[edit]

used in paleoceanography

Role of microorganisms
[edit]"Microbial communities are key drivers of marine biogeochemistry. Our understanding of the incredible complexity and diversity of natural microbial populations has been greatly enhanced by the advent of cultivation-independent techniques for sequencing DNA directly from an environmental sample. Despite progress in describing the complexity of these natural systems, many gaps remain in our understanding of the distribution of genes and organisms in the oceans as well as the selective forces that structure community composition and distribution across space and time."[30]
viruses
[edit]
"Algal viruses are considered to be key players in structuring microbial communities and biogeochemical cycles due to their abundance and diversity within aquatic systems. Their high reproduction rates and short generation times make them extremely successful, often with immediate and strong effects for their hosts and thus in biological and abiotic environments. There are, however, conditions that decrease their reproduction rates and make them unsuccessful with no or little immediate effects. Here, we review the factors that lower viral success and divide them into intrinsic—when they are related to the life cycle traits of the virus—and extrinsic factors—when they are external to the virus and related to their environment. Identifying whether and how algal viruses adapt to disadvantageous conditions will allow us to better understand their role in aquatic systems. We propose important research directions such as experimental evolution or the resurrection of extinct viruses to disentangle the conditions that make them unsuccessful and the effects these have on their surroundings."[31]
"Viruses are the most abundant life forms on Earth, with an estimated 1031 total viruses globally. The majority of these viruses infect microbes, whether bacteria, archaea or microeukaryotes. Given the importance of microbes in driving global biogeochemical cycles, it would seem, based on numerical abundances alone, that viruses also play an important role in the global cycling of carbon and nutrients. However, the importance of viruses in controlling host populations and ecosystem functions, such as the regeneration, storage and export of carbon and other nutrients, remains unresolved. Here, we report on advances in the study of ecological effects of viruses of microbes. In doing so, we focus on an area of increasing importance: the role that ocean viruses play in shaping microbial population sizes as well as in regenerating carbon and other nutrients... The study of viruses of microbes has undergone a revolution in the past 20 years. What has been termed "The Third Age of Phage"[32] can be viewed more broadly as a renewal of interest in environmental viruses, and in particular in viruses of microbes [2-11]. This renewal has been spurred by advances in direct-imaging methods, sequencing technologies and bioinformatics that have revealed a previously unknown world of viral diversity in natural environments [12]. The study of viruses in the oceans has been key to many discoveries: from the finding that viruses have evolved novel lineages of key photosynthetic genes (as in cyanophages infecting Synechococcus and Prochlorococcus [13-17]) to the identification of many novel viral families that defy much of the conventional wisdom about viral life history (e.g. the discovery of “giant” algal-infecting viruses [18,19] and even viruses that exploit other viruses [20]). Altogether, it is well established that a diverse reservoir of viruses infect and lyse bacteria, archaea and microeukaryotes at the base of the ocean's food web [2,21]. Despite the growing literature on viruses of microbes, we remain relatively poorly informed concerning the dynamic, ecological effects of virus infection of marine microbes. In this report, we focus on the functional role of viruses within oceanic waters. In doing so, we highlight knowns, unknowns and challenges in three areas: (i) the role of ocean viruses in shaping microbial community composition; (ii) the role of ocean viruses in determining carbon and nutrient availability; (iii) modelling efforts to link viral infection of microbes with ecosystem-scale consequences."[33]
"In marine environments, virus-mediated lysis of host cells leads to the release of cellular carbon and nutrients and is hypothesized to be a major driver of carbon recycling on a global scale. However, efforts to characterize the effects of viruses on nutrient cycles have overlooked the geochemical potential of the virus particles themselves, particularly with respect to their phosphorus content. In this Analysis article, we use a biophysical scaling model of intact virus particles that has been validated using sequence and structural information to quantify differences in the elemental stoichiometry of marine viruses compared with their microbial hosts. By extrapolating particle-scale estimates to the ecosystem scale, we propose that, under certain circumstances, marine virus populations could make an important contribution to the reservoir and cycling of oceanic phosphorus."[34]
"Marine viruses have important roles in microbial mortality, gene transfer, metabolic reprogramming and biogeochemical cycling. In this Review, we discuss recent technological advances in marine virology including the use of near-quantitative, reproducible metagenomics for large-scale investigation of viral communities and the emergence of gene-based viral ecology. We also describe the reprogramming of microbially driven processes by viral metabolic genes, the identification of novel viruses using cultivation-dependent and cultivation-independent tools, and the potential for modelling studies to provide a framework for studying virus–host interactions. These transformative advances have set a rapid pace in exploring and predicting how marine viruses manipulate and respond to their environment."[35]
"Viruses numerically dominate our oceans; however, we have only just begun to document the diversity, host range and infection dynamics of marine viruses, as well as the subsequent effects of infection on both host cell metabolism and oceanic biogeochemistry. Bacteriophages (that is, phages: viruses that infect bacteria) are highly abundant and are known to play critical roles in bacterial mortality, biogeochemical cycling and horizontal gene transfer. This Review Article summarizes current knowledge of marine viral ecology and highlights the importance of phage particles to the dissolved organic matter pool, as well as the complex interactions between phages and their bacterial hosts. We emphasize the newly recognized roles of phages as puppet masters of their bacterial hosts, where phages are capable of altering the metabolism of infected bacteria through the expression of auxiliary metabolic genes and the redirection of host gene expression patterns. Finally, we propose the ‘royal family model’ as a hypothesis to describe successional patterns of bacteria and phages over time in marine systems, where despite high richness and significant seasonal differences, only a small number of phages appear to continually dominate a given marine ecosystem. Although further testing is required, this model provides a framework for assessing the specificity and ecological consequences of phage–host dynamics."[36]
- Middelboe, M. and Brussaard, C.P. (2017) "Marine viruses: Key players in marine ecosystems", Viruses, 9(10): 302;. doi:10.3390/v9100302.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - See Special Issue "Marine Viruses 2016"
"Viruses are a key component of marine ecosystems, but the assessment of their global role in regulating microbial communities and the flux of carbon is precluded by a paucity of data, particularly in the deep ocean. We assessed patterns in viral abundance and production and the role of viral lysis as a driver of prokaryote mortality, from surface to bathypelagic layers, across the tropical and subtropical oceans. Viral abundance showed significant differences between oceans in the epipelagic and mesopelagic, but not in the bathypelagic, and decreased with depth, with an average power-law scaling exponent of −1.03 km−1 from an average of 7.76 × 106 viruses ml−1 in the epipelagic to 0.62 × 106 viruses ml−1 in the bathypelagic layer with an average integrated (0 to 4000 m) viral stock of about 0.004 to 0.044 g C m−2, half of which is found below 775 m. Lysogenic viral production was higher than lytic viral production in surface waters, whereas the opposite was found in the bathypelagic, where prokaryotic mortality due to viruses was estimated to be 60 times higher than grazing. Free viruses had turnover times of 0.1 days in the bathypelagic, revealing that viruses in the bathypelagic are highly dynamic. On the basis of the rates of lysed prokaryotic cells, we estimated that viruses release 145 Gt C year−1 in the global tropical and subtropical oceans. The active viral processes reported here demonstrate the importance of viruses in the production of dissolved organic carbon in the dark ocean, a major pathway in carbon cycling."[37]
"The encounter and capture of bacteria and phytoplankton by microbial predators and parasites is fundamental to marine ecosystem organization and activity. Here, we combined classic biophysical models with published laboratory measurements to infer functional traits, including encounter kernel and capture efficiency, for a wide range of marine viruses and microzooplankton grazers. Despite virus particles being orders of magnitude smaller than microzooplankton grazers, virus encounter kernels and adsorption rates were in many cases comparable in magnitude to grazer encounter kernel and clearance, pointing to Brownian motion as a highly effective method of transport for viruses. Inferred virus adsorption efficiency covered many orders of magnitude, but the median virus adsorption efficiency was between 5 and 25% depending on the assumed host swimming speed. Uncertainty on predator detection area and swimming speed prevented robust inference of grazer capture efficiency, but sensitivity analysis was used to identify bounds on unconstrained processes. These results provide a common functional trait framework for understanding marine host-virus and predator-prey interactions, and highlight the value of theory for interpreting measured life-history traits."[38]
"Viruses are by far the most abundant entities in marine environments, and are mainly phages that infect bacteria and archaea, which also are a significant component of marine ecosystem and a major force behind marine biogeochemical cycles. As a major source of mortality, viral lysis can release highly labile cellular components, both organic matters and inorganic nutrients, regulating the metabolism of its hosts and influencing the biogeochemical cycles. During infection, viruses could hijack the metabolic system of hosts for its own propagation, thereby changing the metabolism and metabolites of host cells. This paper summarized the effects of viruses on the metabolism of marine bacterioplankton at both the cellular and community level, and its influence on the cycling of ocean elements. Then, the potential impact of environmental factors was assessed on the influence of viruses upon bacterial metabolism. This paper will contribute to a comprehensive understanding of the role of microbes within marine biogeochemical cycles."[39]
bacteria
[edit]"Little is still known of the impacts of protist grazing on bacterioplankton communities in the dark ocean. Furthermore, the accuracy of assessments of in situ microbial activities, including protist grazing, can be affected by sampling artifacts introduced during sample retrieval and downstream manipulations. Potential artifacts may be increased when working with deep-sea samples or samples from chemically unique water columns such as oxygen minimum zones (OMZs). OMZs are oxygen-depleted regions in the ocean, where oxygen concentrations can drop to <20 μM. These regions are typically located near eastern boundary upwelling systems and currently occur in waters occupying below about 8% of total ocean surface area, representing ~1% of the ocean's volume. OMZs have a profound impact not only on the distribution of marine Metazoa, but also on the composition and activities of microbial communities at the base of marine food webs. Here we present an overview of current knowledge of protist phagotrophy below the photic zone, emphasizing studies of oxygen-depleted waters and presenting results of the first attempt to implement new technology for conducting these incubation studies completely in situ (the Microbial Sampling- Submersible Incubation Device, MS-SID). We performed 24-h incubation experiments in the Eastern Tropical South Pacific (ETSP) OMZ. This preliminary study shows that up to 28% of bacterial biomass may be consumed by protists in waters where oxygen concentrations were down to ~4.8 μM and up to 13% at a station with nitrite accumulation where oxygen concentrations were undetectable. Results also show that shipboard measurements of grazing rates were lower than rates measured from the same water using the MS-SID, suggesting that in situ experiments help to minimize artifacts that may be introduced when conducting incubation studies using waters collected from below the photic zone, particularly from oxygen-depleted regions of the water column."[40]
"Prochlorococcus and SAR11 are among the smallest and most abundant organisms on Earth. With a combined global population of about 2.7 × 1028 cells, they numerically dominate bacterioplankton communities in oligotrophic ocean gyres and yet they have never been grown together in vitro. Here we describe co-cultures of Prochlorococcus and SAR11 isolates representing both high- and low-light adapted clades. We examined: (1) the influence of Prochlorococcus on the growth of SAR11 and vice-versa, (2) whether Prochlorococcus can meet specific nutrient requirements of SAR11, and (3) how co-culture dynamics vary when Prochlorococcus is grown with SAR11 compared with sympatric copiotrophic bacteria. SAR11 grew 15–70% faster in co-culture with Prochlorococcus, while the growth of the latter was unaffected. When Prochlorococcus populations entered stationary phase, this commensal relationship rapidly became amensal, as SAR11 abundances decreased dramatically. In parallel experiments with copiotrophic bacteria; however, the heterotrophic partner increased in abundance as Prochlorococcus densities leveled off. The presence of Prochlorococcus was able to meet SAR11’s central requirement for organic carbon, but not reduced sulfur. Prochlorococcus strain MIT9313, but not MED4, could meet the unique glycine requirement of SAR11, which could be due to the production and release of glycine betaine by MIT9313, as supported by comparative genomic evidence. Our findings also suggest, but do not confirm, that Prochlorococcus MIT9313 may compete with SAR11 for the uptake of 3-dimethylsulfoniopropionate (DMSP). To give our results an ecological context, we assessed the relative contribution of Prochlorococcus and SAR11 genome equivalents to those of identifiable bacteria and archaea in over 800 marine metagenomes. At many locations, more than half of the identifiable genome equivalents in the euphotic zone belonged to Prochlorococcus and SAR11 – highlighting the biogeochemical potential of these two groups."[41]
"As the smallest and most abundant primary producer in the oceans, the cyanobacterium Prochlorococcus is of interest to diverse branches of science. For the past 30 years, research on this minimal phototroph has led to a growing understanding of biological organization across multiple scales, from the genome to the global ocean ecosystem. Progress in understanding drivers of its diversity and ecology, as well as molecular mechanisms underpinning its streamlined simplicity, has been hampered by the inability to manipulate these cells genetically. Multiple attempts have been made to develop an efficient genetic transformation method for Prochlorococcus over the years; all have been unsuccessful to date, despite some success with their close relative, Synechococcus... Prochlorococcus is the most abundant cyanobacterium worldwide, dominating vast regions of the global oceans [19]. It may account for up to 50 % of the chlorophyll in oligotrophic ocean regions [20, 21] and is responsible for around 8.5 % of global ocean primary productivity [19]. Our understanding of Prochlorococcus biology and ecology has been greatly facilitated by the rise of metagenomic, metatranscriptomic, and metaproteomic studies of ocean samples over the past decade [22–26]. Because of its high relative abundance, it often dominates databases derived from surface microbial communities in the oceans [27], and has emerged as one of the best-described marine microorganisms with more than one thousand complete or nearly complete genomes available [28–30]. Prochlorococcus is a perfect example of the imbalance between the availability of genomic data and the dearth of genetic tools. Prochlorococcus’ numerical dominance in oligotrophic oceans is attributed to its small size (hence high surface/volume ratio), which enhances its ability to compete for limiting nutrients [31], and its vast genomic diversity [32–37], which expands the niche dimensions of the Prochlorococcus meta-population [28, 38, 39]."[42]
archaea
[edit]"Archaea are ubiquitous and abundant members of the marine plankton. Once thought of as rare organisms found in exotic extremes of temperature, pressure, or salinity, archaea are now known in nearly every marine environment. Though frequently referred to collectively, the planktonic archaea actually comprise four major phylogenetic groups, each with its own distinct physiology and ecology. Only one group—the marine Thaumarchaeota—has cultivated representatives, making marine archaea an attractive focus point for the latest developments in cultivation-independent molecular methods. Here, we review the ecology, physiology, and biogeochemical impact of the four archaeal groups using recent insights from cultures and large-scale environmental sequencing studies. We highlight key gaps in our knowledge about the ecological roles of marine archaea in carbon flow and food web interactions. We emphasize the incredible uncultivated diversity within each of the four groups, suggesting there is much more to be done."[43]
"Archaea constitute a considerable fraction of the microbial biomass on Earth. Like Bacteria they have evolved a variety of energy metabolisms using organic and/or inorganic electron donors and acceptors, and many of them are able to fix carbon from inorganic sources. Archaea thus play crucial roles in the Earth's global geochemical cycles and influence greenhouse gas emissions. Methanogenesis and anaerobic methane oxidation are important steps in the carbon cycle; both are performed exclusively by anaerobic archaea. Oxidation of ammonia to nitrite is performed by Thaumarchaeota. They represent the only archaeal group that resides in large numbers in the global aerobic terrestrial and marine environments on Earth. Sulfur-dependent archaea are confined mostly to hot environments, but metal leaching by acidophiles and reduction of sulfate by anaerobic, nonthermophilic methane oxidizers have a potential impact on the environment. The metabolisms of a large number of archaea, in particular those dominating the subsurface, remain to be explored."[44]
protists
[edit]"Protists are microscopic eukaryotic microbes that are ubiquitous, diverse, and major participants in oceanic food webs and in marine biogeochemical cycles. The study and characterization of protists has a long and distinguished tradition. Even with this history, the extraordinary species diversity and variety of interactions of protists in the sea are only now being fully appreciated. Figure 1 shows representative examples of marine protists, and of methods used to visualize these microbes. Protists can be autotrophic or heterotrophic. The former, also called microscopic algae, contain chloroplasts, thrive by photosynthesis, and are at the base of all oceanic food webs, with the exception of deep-sea chemosynthetic ecosystems. There is a general trend for > 20-µm sized phytoplankton (microplankton), such as diatoms and dinoflagellates, to dominate episodically in coastal waters, while in the open ocean 2–20-µm sized cells (nanoplankton), such as coccolithophorids, sporadically form massive blooms that can be seen from space. However, during nonbloom seasons, even smaller cells (picoplankton—prokaryotes and eukaryotes less than a few micrometers in size) typically dominate phytoplankton biomass and production (Li, 2002; Worden et al., 2004). Some genera of marine picoalgae can “bloom” to very high concentrations (> 105 cells ml-1), Ostreococcus (Countway and Caron, 2006), while others are ubiquitous, such as Micromonas, which is found from arctic to tropical waters (Not et al., 2004; Worden, 2006)."[45]
diatoms
[edit]dinoflagellates
[edit]algae
[edit]coccolithophores
[edit]radiolarians
[edit]foraminiferans
[edit]mixotrophs
[edit]planktonic fungus
[edit]"Fungal contributions to ecosystem processes are well documented for terrestrial systems yet oceans, which account for most of the Earth’s surface, have remained poorly explored with regards to organisms in this kingdom. Here, we demonstrate that, although in low relative abundance (i.e., fungal reads made up 1.4–2.9% of the metagenomes), fungi contribute to both phylogenetic and functional microbial diversity with a conserved fungal presence in global marine samples. Universally distributed taxa and functions implicate them in complex carbon and fatty acid metabolism, with depth stratification along pelagic zones. Functional differences in observed genes between epipelagic and mesopelagic waters indicate changes in UV protection, shift to carbohydrate limited diets, as well as alternative energy sources. Metagenomic data also provided evidence for a latitudinal gradient in fungal diversity linked to temperature shifts. Our results suggest that fungi contribute to multiple biogeochemical cycles in the pelagic ocean, and could be integral for ecosystem functioning through provision of key nutrients."[46]
"Growing interest in understanding the relevance of marine fungi to food webs, biogeochemical cycling, and biological patterns necessitates establishing a context for interpreting future findings. To help establish this context, we summarize the diversity of cultured and observed marine planktonic fungi from across the world. While exploring this diversity, we discovered that only half of the known marine fungal species have a publicly available DNA locus, which we hypothesize will likely hinder accurate high-throughput sequencing classification in the future, as it does currently. Still, we reprocessed >600 high-throughput datasets and analyzed 4.9 × 109 sequences (4.8 × 109 shotgun metagenomic reads and 1.0 × 108 amplicon sequences) and found that every fungal phylum is represented in the global marine planktonic mycobiome; however, this mycobiome is generally predominated by three phyla: the Ascomycota, Basidiomycota, and Chytridiomycota. We hypothesize that these three clades are the most abundant due to a combination of evolutionary histories, as well as physical processes that aid in their dispersal. We found that environments with atypical salinity regimes (>5 standard deviations from the global mean: Red Sea, Baltic Sea, sea ice) hosted higher proportions of the Chytridiomycota, relative to open oceans that are dominated by Dikarya. The Baltic Sea and Mediterranean Sea had the highest fungal richness of all areas explored. An analysis of similarity identified significant differences between oceanographic regions. There were no latitudinal gradients of marine fungal richness and diversity observed. As more high-throughput sequencing data become available, expanding the collection of reference loci and genomes will be essential to understanding the ecology of marine fungi."[47]
plankton
[edit]phytoplankton
[edit]
"Carbon, nitrogen and phosphorus are essential elements required for all life on Earth. In the marine environment, dissolved inorganic carbon, nitrogen and phosphorus are utilized during phytoplankton growth to form organic material, which is respired and remineralized back to inorganic forms by the activity of bacteria, Archaea and zooplankton. The net result of the photosynthesis, calcification and respiration of marine plankton is the uptake of carbon dioxide (CO2) from the atmosphere, its sequestration to the deep ocean as organic and inorganic carbon and its availability to fuel all fish and shellfish production. The cycling of carbon by marine plankton is inextricably linked to that of nitrogen and phosphorus; thus marine plankton mediate climate through influencing the atmospheric concentration of not only CO2, but also nitrous oxide (N2O). Increasing anthropogenically derived atmospheric CO2 concentrations impact plankton mediated biogeochemical cycles through increasing seawater temperature and dissolution of CO2, leading to changes in water column mixing, availability of light and nutrients, decreasing dissolved oxygen and changing carbonate chemistry. This chapter describes how the activity of phytoplankton, bacteria and Archaea drive the marine biogeochemical cycles of carbon, nitrogen and phosphorus, and how climate driven changes in plankton abundance and community composition are influencing these biogeochemical cycles in the North Atlantic Ocean and adjacent seas."[56]
zooplankton
[edit]"Exploring climate and anthropogenic impacts on marine ecosystems requires an understanding of how trophic components interact. However, integrative end-to-end ecosystem studies (experimental and/or modelling) are rare. Experimental investigations often concentrate on a particular group or individual species within a trophic level, while tropho-dynamic field studies typically employ either a bottom-up approach concentrating on the phytoplankton community or a top-down approach concentrating on the fish community. Likewise the emphasis within modelling studies is usually placed upon phytoplankton-dominated biogeochemistry or on aspects of fisheries regulation. In consequence the roles of zooplankton communities (protists and metazoans) linking phytoplankton and fish communities are typically under-represented if not (especially in fisheries models) ignored. Where represented in ecosystem models, zooplankton are usually incorporated in an extremely simplistic fashion, using empirical descriptions merging various interacting physiological functions governing zooplankton growth and development, and thence ignoring physiological feedback mechanisms. Here we demonstrate, within a modelled plankton food-web system, how trophic dynamics are sensitive to small changes in parameter values describing zooplankton vital rates and thus the importance of using appropriate zooplankton descriptors. Through a comprehensive review, we reveal the mismatch between empirical understanding and modelling activities identifying important issues that warrant further experimental and modelling investigation. These include: food selectivity, kinetics of prey consumption and interactions with assimilation and growth, form of voided material, mortality rates at different age-stages relative to prior nutrient history. In particular there is a need for dynamic data series in which predator and prey of known nutrient history are studied interacting under varied pH and temperature regimes."[57]
- Everett, J.D., Baird, M.E., Buchanan, P., Bulman, C., Davies, C., Downie, R., Griffiths, C., Heneghan, R., Kloser, R.J., Laiolo, L. and Lara-Lopez, A. (2017) "Modeling what we sample and sampling what we model: challenges for zooplankton model assessment". Frontiers in Marine Science, 4: 77. doi:10.3389/fmars.2017.00077.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Classic food web and the microbial loop
[edit]
solid arrows: inputs; dashed arrows: outputs; blue arrows: mixing; red and green arrows: exchange between the mixed surface layer and lower water [57]
Azam's paper...
- Azam, F., T. Fenchel, J.G. Field, J.S. Gray, L.A. Meyer-Reil, and F. Thingstad (1983) [www.jstor.org/stable/24814647 "The ecological role of water-column microbes in the sea"]. Marine Ecology Progress Series, 10: 257-263.
On the "classic" food web (though he doesn't use the word "classic") and the microbial loop.
"Prior to the 1980s, the structure of the ecosystem in the pelagic marine waters was typically described through what is now termed the “classical” food web (Steele, 1974, Cushing, 1975). Within this structure, primary production is attributed to photoautotrophic phytoplankton. These phytoplankton are then consumed by the “herbivorous” zooplankton (i.e., primary consumers) which are in turn ingested by carnivorous zooplankton and pelagic fish, which then serve as food for larger fish. Despite some earlier suggestions to modify this classic food web structure (e.g., Pomeroy, 1974), it was not until the early 1980s that the importance of microbial production gained recognition (Williams, 1981, Fenchel, 1982), and the planktonic food web concept was broadened towards a more integrated view (the microbial food web). In this new defined structure phytoplankton as well as bacteria are consumed by protozoan grazers (Sherr and Sherr, 1994, Calbet, 2008), thus providing an additional food source for copepods and higher trophic levels. Following such studies, Azam et al. (1983) proposed the “microbial loop” as an addition to the food web, within which dissolved organic carbon (DOC) is reincorporated into the food web, mediated by microbial activity."[57]
- - mixotrophic
Gifford's paper:
- Gifford, D.J. (1991) "The protozoan‐metazoan trophic link in pelagic ecosystems". The Journal of Protozoology, 38(1): 81–86. doi:10.1111/j.1550-7408.1991.tb04806.x.
"The recognition of the importance of the microbial loop led to the “link-sink” debate (Gifford, 1991), questioning whether the activity of the protozoan grazers served as a “link” between the microbial loop and the classical food chain (Sanders and Porter, 1987), or as a “sink” for carbon (Ducklow et al., 1986). Various field studies, experimental results and modelling efforts have subsequently shown microzooplankton to be a link between the classical and microbial food webs in marine as well as fresh water bodies thus acting as conduits of energy and nutrients between the microbial level and higher trophic levels (Suttle et al., 1986, Frost, 1987, Cushing, 1995, Calbet and Saiz, 2005). Additionally, based on stoichiometric and biochemical grounds, microzooplankton, rather than phytoplankton, could be expected to be better prey for mesozooplankton (Klein Breteler et al., 1999, Broglio et al., 2003, Mitra and Flynn, 2005). The latest twist to this is the concept that much of the plankton community currently split between either phototrophic phytoplankton or heterotrophic microzooplankton should be recognised as mixotrophic (Flynn et al., 2013, Mitra et al., 2014)".[57]
- - mathematical descriptions
"Today, the construction, testing and deployment of mathematical descriptions of plankton dynamics are central planks in marine ecology and climate change research. Many of these studies are based on the classic ecosystem model of Fasham et al. (1990), or variations on that theme. However, while over the last half century our understanding of aquatic ecology has undergone a substantial change, models portraying these systems have not developed in line with field and laboratory observations. Model structure and complexity has not typically changed in ecosystem models to reflect improvements in our understanding of biological complexity with its attendant feedback mechanisms (Mitra and Davis, 2010, Rose et al., 2010). The dramatic increase in model complexity over this period has been almost wholly focussed on the phytoplankton–nutrient link, with regard to variables, processes and parameters. Very little, by comparison, has been done with the Z component, quite often employing only 2 classes (e.g., 78 P boxes vs. 2 Z boxes in Follows et al., 2007). Despite the plethora of mechanistic zooplankton models which have been developed over the past two decades (e.g., Carlotti and Hirche, 1997, Carlotti and Wolf, 1998, Mitra, 2006, Mitra and Flynn, 2007, Flynn and Irigoien, 2009), the Z-boxes within ecosystem models are still biologically extremely simplistic with little or no differences in the physiological descriptions between the different Z-boxes. This is despite the manifest difference in the ecophysiology of the protist microzooplankton and the metazoan zooplankton. Increased complexity has usually been in numerical rather than detailed structural complexity; for example, 1-box representing the entire zooplankton (Z) community vs. 3-boxes representing different zooplankton functional types (e.g., Franks, 2002 vs. Blackford et al., 2004)."[57]
- - top down/bottom up
"The zooplankton community has thus been typically side-lined within ecosystem studies, not receiving the same level of importance as the phytoplankton and fish communities. Within biogeochemical models, zooplankton represent the top trophic level acting as a closure function, while within many fisheries models, zooplankton form the bottom level (see reviews by Plagányi, 2007, Carlotti and Poggiale, 2010, Fulton, 2010). However, there is a growing recognition of the need to bring together these two strands of research (biogeochemical and fisheries) through development of end-to-end ecosystem models combining physicochemical oceanographic descriptors with the biology of all trophic levels from microbes to higher-trophic-level, including humans, in a single modelling framework (Mitra and Davis, 2010, Rose et al., 2010). Fig. 1 presents a conceptual model of such an end-to-end food web ecosystem. The zooplankton community (Z) acts as the conduit for the transfer of energy and material from the primary producers to the higher trophic levels and has a pivotal role in recycling and export of nutrients. Thus the zooplankton community is the critical link between biogeochemistry and fisheries (Carlotti and Poggiale, 2010, Mitra and Davis, 2010).
inverted pyramids
[edit]




microzooplankton
[edit]microbenthos
[edit]mineral dust
[edit]Nutrients in the dust
"One of the major sources of essential nutrients to the ocean is mineral dust, which can deposit nitrogen, phosphorus, and iron, a biologically-important trace metal, into the open ocean, making mineral dust deposition an important factor controlling phytoplankton growth. The Saharan and Gobi Deserts are major sources of mineral dusts, which can be transported from source areas over thousands of kilometers. Climate change models forecast increased levels of dust deposition on the open ocean, and so the study here set out to understand how dust additions impact diatom growth."[8]
trace elements
[edit]"Trace elements serve important roles as regulators of ocean processes including marine ecosystem dynamics and carbon cycling. The role of iron, for instance, is well known as a limiting micronutrient in the surface ocean. Several other trace elements also play crucial roles in ecosystem function and their supply therefore controls the structure, and possibly the productivity, of marine ecosystems. Understanding the biogeochemical cycling of these micronutrients requires knowledge of their diverse sources and sinks, as well as their transport and chemical form in the ocean. Much of what is known about past ocean conditions, and therefore about the processes driving global climate change, is derived from trace-element and isotope patterns recorded in marine deposits. Reading the geochemical information archived in marine sediments informs us about past changes in fundamental ocean conditions such as temperature, salinity, pH, carbon chemistry, ocean circulation and biological productivity. These records provide our principal source of information about the ocean’s role in past climate change. Understanding this role offers unique insights into the future consequences of global change. The cycle of many trace elements and isotopes has been significantly impacted by human activity. Some of these are harmful to the natural and human environment due to their toxicity and/or radioactivity. Understanding the processes that control the transport and fate of these contaminants is an important aspect of protecting the ocean environment. Such understanding requires accurate knowledge of the natural biogeochemical cycling of these elements so that changes due to human activity can be put in context".[59]
Biomass composition
[edit]"Obtaining a quantitative global picture of life in the great expanses of the oceans is a challenging task. By integrating data from across existing literature, we provide a comprehensive view of the distribution of marine biomass between taxonomic groups, modes of life, and habitats."[60]


Modeling biological oceanography
[edit]
The example model is the Bering Ecosystem Study Nutrient-Phytoplankton-Zooplankton model (BESTNPZ)
- Marine biogeochemical models
- Marine food web models
"Nearly 100 years ago, Alfred Lotka published two short but insightful papers[65][66] describing how ecosystems may organize. Principally, Lotka argued that ecosystems will grow in size and that their cycles will spin faster via predation and nutrient recycling so as to capture all available energy, and that evolution and natural selection are the mechanisms by which this occurs and progresses. Lotka's ideas have often been associated with the maximum power principle, but they are more consistent with recent developments in nonequilibrium thermodynamics, which assert that complex systems will organize toward maximum entropy production (MEP).[67] [68] In this review, we explore Lotka's hypothesis within the context of the MEP principle,[69][70][71] as well as how this principle can be used to improve aquatic biogeochemistry models. We need to develop the equivalent of a climate model, as opposed to a weather model, to understand marine biogeochemistry on longer timescales, and adoption of the MEP principle can help create such models."[72][73]
"In current process-based biogeochemical models, mathematical expressions derived from available theoretical or empirical relationships are used to define the interactions between numerous state variables. The parameterization of these approximate mathematical expressions relies mainly on maximizing agreement between simulations and observations. These models are highly nonlinear, location-specific, and challenging to scale or apply in a different environment. The Maximum Energy Production (MEP) modelling approach presented here is based on the hypothesis that living systems evolve and organize to maximize free energy dissipation over the greatest possible spatial and temporal scales. In this study, we used a trait-based global ocean modeling system (MITgcm Darwin) to calculate entropy fluxes among different biotic constituents in the euphotic zone. Reaction free energies associated with different metabolic functions in a biological system, such as growth, nutrient uptake, nitrogen fixation, and denitrification, were calculated using simplified stoichiometric equations and reaction rates derived from the model for simple and complex food web structures. The preliminary results suggest that maximum entropy production can be used as a criterion to select food web configurations that may better represent natural marine ecosystems."[74]
FROM Alfred J. Lotka... His earlier work was centered on energetics and applications of thermodynamics in life sciences. Lotka proposed the theory that the Darwinian concept of natural selection could be quantified as a physical law. The law that he proposed was that the selective principle of evolution was one which favoured the maximum useful energy flow transformation. The general systems ecologist Howard T. Odum later applied Lotka's proposal as a central guiding feature of his work in ecosystem ecology. Odum called Lotka's law the maximum power principle.
For the drama of life is like a puppet show in which stage, scenery, actors and all are made of the same stuff... and if we would catch the spirit of the piece, our attention must not be absorbed in the characters alone, but must be extended also to the scene, of which they are born, on which they play their part, and with which, in a little while, they merge again
— Alfred Lotka (1925)
- For the drama of life is like a puppet show in which stage, scenery, actors and all are made of the same stuff... and if we would catch the spirit of the piece, our attention must not be absorbed in the characters alone, but must be extended also to the scene, of which they are born, on which they play their part, and with which, in a little while, they merge again – Alfred Lotka (1925)
- What did Lotka really say?[75]
- How reliable are ocean models?
- Raäisaänen, J. (2007) "How reliable are climate models?". Tellus A: Dynamic Meteorology and Oceanography, 59(1): 2–29. doi:10.1111/j.1600-0870.2006.00211.x.
- How reliable are climate models?, Skeptical Science.
- Smith, Ryan & Kelly, Jonathan & Nazarzadeh, Kimia & Sukhatme, Gaurav. (2013). An Investigation on the Accuracy of Regional Ocean Models Through Field Trials. Proceedings - IEEE International Conference on Robotics and Automation. 10.1109/ICRA.2013.6631057.
- Ocean general circulation model
Marine sediments
[edit]
Fish populations are linked to ocean biogeochemistry by their reliance on primary production for food, and dissolved oxygen to breathe. It is also possible that marine fish modify biogeochemical dynamics, as do freshwater fish, through top-down trophic cascades, but there has been relatively little consideration of this possibility. This lack of consideration may reflect a lack of importance; alternatively, it may simply reflect the lack of appropriate observations with which to constrain such relationships. Here, we draw attention to the potential use of marine sediments as long-term simultaneous monitors of both fish abundance and marine biogeochemical dynamics. We compile published sediment proxy records of fish abundance from the west coasts of the Americas, and compare them with biogeochemical climatic signals, we find a small number of statistically significant relationships between fish debris and biogeochemical variables, at least some of which are likely to reflect causal relationships. Considering total organic carbon(TOC), the most commonly-measured biogeochemical variable, some positive correlations with fish abundance are found, consistent with bottom-up control of fish abundance by primary production, or a planktivore-herbivore-phytoplankton trophic cascade. Negative correlations are also found, which could reflect sedimentary processes, the influence of upwelling-driven oxygen and nutrient dynamics on primary production and fish populations, and/or impacts of fish stocks on carbon fluxes by altering the recycling of carbon within the water column. Although the number of available measurements is too small to draw strong conclusions, the results point to plausible cases of bottom-up forcing, trophic cascades, and influence of dissolved oxygen concentrations on fish habitat.[76]
Benthic foraminifera
[edit]
- Schmiedl, G. (2019) "Use of Foraminifera in Climate Science". In: Oxford Research Encyclopedia of Climate Science: Past Climates. doi:10.1093/acrefore/9780190228620.013.735.
Anthropogenic changes
[edit]"The prokaryotic and eukaryotic microorganisms that drive the pelagic ocean's biogeochemical cycles are currently facing an unprecedented set of comprehensive anthropogenic changes. Nearly every important control on marine microbial physiology is currently in flux, including seawater pH, pCO2, temperature, redox chemistry, irradiance and nutrient availability. Here, we examine how microorganisms with key roles in the ocean carbon and nitrogen cycles may respond to these changes in the Earth's largest ecosystem. Some functional groups such as nitrogen-fixing cyanobacteria and denitrifiers may be net beneficiaries of these changes, while others such as calcifiers and nitrifiers may be negatively impacted. Other groups, such as heterotrophic bacteria, may be relatively resilient to changing conditions. The challenge for marine microbiologists will be to predict how these divergent future responses of marine microorganisms to complex multiple variable interactions will be expressed through changing biogeography, community structure and adaptive evolution, and ultimately through large-scale alterations of the ocean's carbon and nutrient cycles."[79]

- megafauna
"In a paper published in Science Advances, an international team of researchers have examined traits of marine megafauna species to better understand the potential ecological consequences of their extinction under different future scenarios. Defined as the largest animals in the oceans, with a body mass that exceeds 45kg, examples include sharks, whales, seals and sea turtles. These species serve key roles in ecosystems, including the consumption of large amounts of biomass, transporting nutrients across habitats, connecting ocean ecosystems, and physically modifying habitats."[83][84]
"Habitat loss is accelerating a global extinction crisis. Conservation requires understanding links between species and habitats. Emerging research is revealing important associations between vegetated coastal wetlands and marine megafauna, such as cetaceans, sea turtles, and sharks. But these links have not been reviewed and the importance of these globally declining habitats is undervalued. Here, we identify associations for 102 marine megafauna species that utilize these habitats, increasing the number of species with associations based on current International Union for the Conservation of Nature (IUCN) species assessments by 59% to 174, accounting for over 13% of all marine megafauna. We conclude that coastal wetlands require greater protection to support marine megafauna, and present a simple, effective framework to improve the inclusion of habitat associations within species assessments."[85]
- references
- ^ Cardini, U., Bednarz, V.N., Foster, R.A. and Wild, C. (2014) "Benthic N2 fixation in coral reefs and the potential effects of human‐induced environmental change". Ecology and evolution, 4(9): 1706–1727. doi:10.1002/ece3.1050
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Adapted from: Gruber, N., and J. N. Galloway (2008) "An Earth‐system perspective of the global nitrogen cycle". Nature, 451:293–296. doi:10.1038/nature06592.
- ^ Raina, J.B. (2018) "The life aquatic at the microscale". mSystems, 3(2): e00150-17. doi:10.1128/mSystems.00150-17.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b c Wadham, J.L., Hawkings, J.R., Tarasov, L., Gregoire, L.J., Spencer, R.G.M., Gutjahr, M., Ridgwell, A. and Kohfeld, K.E. (2019) "Ice sheets matter for the global carbon cycle". Nature communications, 10(1): 1–17. doi:10.1038/s41467-019-11394-4.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Sarmiento, Jorge Louis (2006). Ocean biogeochemical dynamics. Gruber, Nicolas. Princeton: Princeton University Press. ISBN 9780691017075. OCLC 60651167.
- ^ Conley, Daniel J. (December 2002). "Terrestrial ecosystems and the global biogeochemical silica cycle". Global Biogeochemical Cycles. 16 (4): 68–1–68–8. Bibcode:2002GBioC..16.1121C. doi:10.1029/2002gb001894. ISSN 0886-6236. S2CID 128672790.
- ^ Drever, James I. (1993). "The effect of land plants on weathering rates of silicate minerals". Geochimica et Cosmochimica Acta. 58 (10): 2325–2332. doi:10.1016/0016-7037(94)90013-2.
- ^ De La Rocha, Christina; Conley, Daniel J. (2017), "The Venerable Silica Cycle", Silica Stories, Springer International Publishing, pp. 157–176, doi:10.1007/978-3-319-54054-2_9, ISBN 9783319540542
- ^ Chadwick, Oliver A.; Ziegler, Karen; Kurtz, Andrew C.; Derry, Louis A. (2005). "Biological control of terrestrial silica cycling and export fluxes to watersheds". Nature. 433 (7027): 728–731. Bibcode:2005Natur.433..728D. doi:10.1038/nature03299. PMID 15716949. S2CID 4421477.
- ^ Fulweiler, Robinson W.; Carey, Joanna C. (2012-12-31). "The Terrestrial Silica Pump". PLOS ONE. 7 (12): e52932. Bibcode:2012PLoSO...752932C. doi:10.1371/journal.pone.0052932. PMC 3534122. PMID 23300825.
- ^ a b c d e Tréguer, Paul J.; De La Rocha, Christina L. (2013-01-03). "The World Ocean Silica Cycle". Annual Review of Marine Science. 5 (1): 477–501. doi:10.1146/annurev-marine-121211-172346. ISSN 1941-1405. PMID 22809182.
- ^ a b Yool, Andrew; Tyrrell, Toby (2003). "Role of diatoms in regulating the ocean's silicon cycle". Global Biogeochemical Cycles. 17 (4): 14.1–14.22. Bibcode:2003GBioC..17.1103Y. CiteSeerX 10.1.1.394.3912. doi:10.1029/2002GB002018. S2CID 16849373.
- ^ DeMaster, David (2002). "The accumulation and cycling of biogenic silica in the Southern Ocean: revisiting the marine silica budget". Deep Sea Research Part II. 49 (16): 3155–3167. Bibcode:2002DSRII..49.3155D. doi:10.1016/S0967-0645(02)00076-0.
- ^ Sutton, Jill N.; Andre, Luc; Cardinal, Damien; Conley, Daniel J.; de Souza, Gregory F.; Dean, Jonathan; Dodd, Justin; Ehlert, Claudia; Ellwood, Michael J. (2018). "A Review of the Stable Isotope Bio-geochemistry of the Global Silicon Cycle and Its Associated Trace Elements". Frontiers in Earth Science. 5. doi:10.3389/feart.2017.00112. ISSN 2296-6463.
- ^ a b c d e Conley, Daniel J. (December 2002). "Terrestrial ecosystems and the global biogeochemical silica cycle". Global Biogeochemical Cycles. 16 (4): 68–1–68-8. doi:10.1029/2002gb001894. ISSN 0886-6236. S2CID 128672790. [verification needed]
- ^ Naveira Garabato, A.C., MacGilchrist, G.A., Brown, P.J., Evans, D.G., Meijers, A.J. and Zika, J.D. (2017) "High-latitude ocean ventilation and its role in Earth's climate transitions". Philosophical Transactions of the Royal Society A: Mathematical, Physical and Engineering Sciences, 375(2102): 20160324. doi:10.1098/rsta.2016.0324.
- ^ Shepherd, J.G., Brewer, P.G., Oschlies, A. and Watson, A.J. (2017) "Ocean ventilation and deoxygenation in a warming world: introduction and overview". Philosophical Transactions of the Royal Society B: Mathematical, Physical and Engineering Sciences,375(2102): doi:10.1098/rsta.2017.0240.
- ^ MacGilchrist, G.A., Marshall, D.P., Johnson, H.L., Lique, C. and Thomas, M. (2017) "Characterizing the chaotic nature of ocean ventilation". Journal of Geophysical Research: Oceans, 122(9): 7577–7594. doi:10.1002/2017JC012875.
- ^ Robinson, C. (2019) "Microbial respiration, the engine of ocean deoxygenation". Frontiers in Marine Science, 5: 533. doi:10.3389/fmars.2018.00533.
- ^ a b Petersen, J.K., Holmer, M., Termansen, M. and Hasler, B. (2019) "Nutrient extraction through bivalves". In: Smaal A., Ferreira J., Grant J., Petersen J., Strand Ø. (eds) Goods and Services of Marine Bivalves, pages 179–208. Springer. ISBN 9783319967769
- ^ Otero, X.L., De La Peña-Lastra, S., Pérez-Alberti, A., Ferreira, T.O. and Huerta-Diaz, M.A. (2018) "Seabird colonies as important global drivers in the nitrogen and phosphorus cycles". Nature communications, 9(1): 1–8. doi:10.1038/s41467-017-02446-8
- ^ Luo, Z., Hu, S. and Chen, D. (2018) "The trends of aquacultural nitrogen budget and its environmental implications in China". Scientific Reports, 8(1): 1–9. doi:10.1038/s41598-018-29214-y
- ^ Basu, S., Gledhill, M., de Beer, D., Matondkar, S.P. and Shaked, Y. (2019) "Colonies of marine cyanobacteria Trichodesmium interact with associated bacteria to acquire iron from dust". Nature:Communications biology, 2(1): 1–8. doi:10.1038/s42003-019-0534-z.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Pajares Moreno, S. and Ramos, R. (2019) "Processes and Microorganisms Involved in the Marine Nitrogen Cycle: Knowledge and Gaps". Frontiers in Marine Science, 6: 739. doi:10.3389/fmars.2019.00739.
- ^ Pajares Moreno, S. and Ramos, R. (2019) "Processes and Microorganisms Involved in the Marine Nitrogen Cycle: Knowledge and Gaps". Frontiers in Marine Science, 6: 739. doi:10.3389/fmars.2019.00739.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Pajares Moreno, S. and Ramos, R. (2019) "Processes and Microorganisms Involved in the Marine Nitrogen Cycle: Knowledge and Gaps". Frontiers in Marine Science, 6: 739. doi:10.3389/fmars.2019.00739.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ SLuo, Z., Hu, S. and Chen, D. (2018) "The trends of aquacultural nitrogen budget and its environmental implications in China". Nature: Science Reports, 8(10877). doi:10.1038/s41598-018-29214-y.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Thomas, E.R., Allen, C.S., Etourneau, J., King, A.C., Severi, M., Winton, V.H.L., Mueller, J., Crosta, X. and Peck, V.L. (2019) "Antarctic sea ice proxies from marine and ice core archives suitable for reconstructing sea ice over the past 2000 years". Geosciences, 9(12): 506. doi:10.3390/geosciences9120506.
- ^ Sallée, J.B. (2018) "Southern ocean warming". Oceanography, 31(2): 52–62. doi:10.5670/oceanog.2018.215.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Biller, S.J., Berube, P.M., Dooley, K., Williams, M., Satinsky, B.M., Hackl, T., Hogle, S.L., Coe, A., Bergauer, K., Bouman, H.A. and Browning, T.J. (2018) "Marine microbial metagenomes sampled across space and time". Scientific data, 5: 180176. doi:10.1038/sdata.2018.176.
- ^ a b Horas, E.L., Theodosiou, L. and Becks, L. (2018) "Why are algal viruses not always successful?", Viruses, 10(9): 474. doi:10.3390/v10090474.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Mann NH (2005) "The third age of phage", PLoS Biology, 3(5): e182. doi:10.1371/journal.pbio.0030182.
- ^ Weitz, Joshua S. and Wilhelm, Steven W. (2013) "An Ocean of Viruses", F1000 Biology Reports, 4: 17. doi:10.3410/B4-17.
- ^ Jover, L.F., Effler, T.C., Buchan, A., Wilhelm, S.W. and Weitz, J.S. (2014) "The elemental composition of virus particles: implications for marine biogeochemical cycles". Nature Reviews Microbiology, 12(7): 519–528. doi:10.1126/sciadv.1602565.
- ^ Brum, J.R. and Sullivan, M.B. (2015) "Rising to the challenge: accelerated pace of discovery transforms marine virology". Nature Reviews Microbiology, 13(3): 147–159. doi:10.1038/nrmicro3404.
- ^ Breitbart, M., Bonnain, C., Malki, K. and Sawaya, N.A. (2018) "Phage puppet masters of the marine microbial realm". Nature Microbiology, 3(7): 754–766. doi:10.1038/s41564-018-0166-y.
- ^ Lara, E., Vaqué, D., Sà, E.L., Boras, J.A., Gomes, A., Borrull, E., Díez-Vives, C., Teira, E., Pernice, M.C., Garcia, F.C. and Forn, I. (2017) "Unveiling the role and life strategies of viruses from the surface to the dark ocean". Science advances, 3(9): p.e1602565. doi:10.1126/sciadv.1602565.
- ^ Talmy, D., Beckett, S.J., Taniguchi, D.A., Zhang, A.B., Weitz, J.S. and Follows, M.J. (2019) "Contrasting controls on microzooplankton grazing and viral infection of microbial prey". Frontiers in Marine Science, 6: 182. doi:10.3389/fmars.2019.00182.
- ^ Longfei, L., Rui, Z., Jie, X. and Nianzhi, J. (2018) "Influence of Virus upon the Marine Bacterial Metabolism and Its Biogeochemical Effects". Advances in Earth Science, 33(3): 225–235. doi:10.11867/j.issn.1001-8166.2018.03.0225.
- ^ Medina, L.E., Taylor, C.D., Pachiadaki, M.G., Henríquez-Castillo, C., Ulloa, O. and Edgcomb, V.P. (2017) "A review of protist grazing below the photic zone emphasizing studies of oxygen-depleted water columns and recent applications of in situ approaches". Frontiers in Marine Science, 4: 105. doi:10.3389/fmars.2017.00105.
- ^ Becker, J.W., Hogle, S.L., Rosendo, K. and Chisholm, S.W. (2019) "Co-culture and biogeography of Prochlorococcus and SAR11". The ISME Journal, 13(6): 1506–1519. doi:10.1038/s41396-019-0365-4.
- ^ Laurenceau, R., Bliem, C., Osburne, M.S., Becker, J.W., Biller, S.J., Cubillos-Ruiz, A. and Chisholm, S.W. (2020) "Toward a genetic system in the marine cyanobacterium Prochlorococcus". Access Microbiology, p.acmi000107. doi:10.1099/acmi.0.000107.
- ^ Santoro, A.E., Richter, R.A. and Dupont, C.L. (2019) "Planktonic marine archaea". Annual review of marine science, 11: 131–158. doi:10.1146/annurev-marine-121916-063141.
- ^ Offre, P., Spang, A. and Schleper, C. (2013) "Archaea in biogeochemical cycles". Annual Review of Microbiology, 67:437–457. doi:10.1146/annurev-micro-092412-155614.
- ^ Sherr, B.F., Sherr, E.B., Caron, D.A., Vaulot, D. and Worden, A.Z. (2007) "Oceanic protists". Oceanography, 20(2): 130–134. doi:10.5670/oceanog.2007.57 (full text)
- ^ Morales, S.E., Biswas, A., Herndl, G.J. and Baltar, F. (2019) "Global structuring of phylogenetic and functional diversity of pelagic fungi by depth and temperature". Frontiers in Marine Science, 6: 131. doi:10.3389/fmars.2019.00131.
- ^ Hassett, B.T., Vonnahme, T.R., Peng, X., Jones, E.G. and Heuzé, C. (2020) "Global diversity and geography of planktonic marine fungi". Botanica Marina, 63(2): 121–139. doi:10.1515/bot-2018-0113.
- ^ a b Brum, J., Schenck, R., and Sullivan, M. (2013) "Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses". ISME J. 7: 1738–1751. doi:10.1038/ismej.2013.67.
- ^ Jover, L. F., Effler, T. C., Buchan, A., Wilhelm, S. W., and Weitz, J. S. (2014) "The elemental composition of virus particles: implications for marine biogeochemical cycles". Nat. Rev. Microbiol. 12: 519–528. doi:10.1038/nrmicro3289.
- ^ Brussaard, C. P. D. (2004) "Viral control of phytoplankton populations - a review". J. Eukaryot. Microbiol. 51: 125–138. doi:10.1111/j.1550-7408.2004.tb00537.x.
- ^ Abergel, C., Legendre, M., and Claverie, J. M. (2015) "The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus". FEMS Microbiol. Rev. 39: 779–796. doi:10.1093/femsre/fuv037.
- ^ Taniguchi, D. A. A., Franks, P. J. S., and Poulin, F. J. (2014) "Planktonic biomass size spectra: an emergent property of size-dependent physiologicalrates, food web dynamics, and nutrient regimes". Mar. Ecol. Prog. Ser. 514: 13–33. doi:10.3354/meps10968.
- ^ DeLong, J. P., Okie, J. G., Moses, M. E., Sibly, R. M., and Brown, J. H. (2010) "Shifts in metabolic scaling, production, and efficiency across major evolutionary transitions of life". Proc. Natl. Acad. Sci. U.S.A. 107: 12941–12945. doi:10.1073/pnas.1007783107.
- ^ Felsenstein, J. (2017) "Phylogenies and the comparative method". Am. Nat. 125: 1–15. doi:10.1111/j.1558-5646.1985.tb00420.x.
- ^ Talmy, D., Beckett, S.J., Taniguchi, D.A., Zhang, A.B., Weitz, J.S. and Follows, M.J. (2019) "Contrasting controls on microzooplankton grazing and viral infection of microbial prey". Frontiers in Marine Science, 6: 182. doi:10.3389/fmars.2019.00182.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Robinson, C. (2017) "Phytoplankton biogeochemical cycles". In: In: Marine Plankton, Oxford University Press. ISBN 9780199233267
- ^ a b c d e Mitra, A., Castellani, C., Gentleman, W.C., Jónasdóttir, S.H., Flynn, K.J., Bode, A., Halsband, C., Kuhn, P., Licandro, P., Agersted, M.D. and Calbet, A. (2014) "Bridging the gap between marine biogeochemical and fisheries sciences; configuring the zooplankton link". Progress in Oceanography, 129: 176–199. doi:10.1016/j.pocean.2014.04.025.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ a b Woodson, C.B., Schramski, J.R. and Joye, S.B. (2018) "A unifying theory for top-heavy ecosystem structure in the ocean". Nature Communications, 9(1): 1-8. doi:10.1038/s41467-017-02450-y.
- ^ SCOR Working Group (2007) "GEOTRACES–An international study of the global marine biogeochemical cycles of trace elements and their isotopes". Geochemistry, 67(2): 85–131. doi:10.1016/j.chemer.2007.02.001.
- ^ Bar-On, Y.M. and Milo, R. (2019) "The Biomass Composition of the Oceans: A Blueprint of Our Blue Planet". Cell, 179(7): 1451–1454. doi:10.1016/j.cell.2019.11.018.
- ^ Heinrichs, M.E., Mori, C. and Dlugosch, L. (2020) "Complex Interactions Between Aquatic Organisms and Their Chemical Environment Elucidated from Different Perspectives". In YOUMARES 9-The Oceans: Our Research, Our Future, pages 279–297, Springer. doi:10.1007/978-3-030-20389-4_15.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Based on: Carlson CA (2002) "Production and removal processes". In: Hansell DA, Carlson CA (eds) Biogeochemistry of marine dissolved organic matter. Academic Press, San Diego, pages 91–151. ISBN 9780123238412.
- ^ Heinrichs, M.E., Mori, C. and Dlugosch, L. (2020) "Complex Interactions Between Aquatic Organisms and Their Chemical Environment Elucidated from Different Perspectives". In YOUMARES 9-The Oceans: Our Research, Our Future, pages 279–297, Springer. doi:10.1007/978-3-030-20389-4_15.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Kearney, K., Hermann, A., Cheng, W., Ortiz, I. and Aydin, K. (2020) "A coupled pelagic–benthic–sympagic biogeochemical model for the Bering Sea: documentation and validation of the BESTNPZ model (v2019. 08.23) within a high-resolution regional ocean model". Geoscientific Model Development, 13(2). doi:10.5194/gmd-13-597-2020.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Lotka A. J. (1922a) "Contribution to the energetics of evolution". Proc Natl Acad Sci USA, 8: pp. 147–51.
- ^ Lotka A. J. (1922b) "Natural selection as a physical principle". Proc Natl Acad Sci USA, 8, pp. 151–54.
- ^ Martyushev, L.M. and Seleznev, V.D. (2006) "Maximum entropy production principle in physics, chemistry and biology". Physics reports, 426(1): 1-45. doi:10.1016/j.physrep.2005.12.001
- ^ Martyushev, L.M. (2013) "Entropy and entropy production: Old misconceptions and new breakthroughs". Entropy, 15(4): 1152–1170. doi:10.3390/e15041152
- ^ Vallino, J.J. (2010) "Ecosystem biogeochemistry considered as a distributed metabolic network ordered by maximum entropy production". Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1545): 1417–1427. doi:10.1098/rstb.2009.0272
- ^ Kleidon, A., Malhi, Y. and Cox, P.M. (2010) "Maximum entropy production in environmental and ecological systems". Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1545): 1297–1302. doi:10.1098/rstb.2010.0018
- ^ Pross, A. and Pascal, R. (2017) "How and why kinetics, thermodynamics, and chemistry induce the logic of biological evolution". Beilstein journal of organic chemistry, 13(1): 665–674. doi:10.3762/bjoc.13.66
- ^ Vallino, J.J. and Algar, C.K. (2016) "The thermodynamics of marine biogeochemical cycles: Lotka revisited", Annual Review of Marine Science, 8: 333–356. doi:10.1146/annurev-marine-010814-015843.
- ^ Vallino, J.J. and Huber, J.A. (2018) "Using maximum entropy production to describe microbial biogeochemistry over time and space in a meromictic pond". Frontiers in Environmental Science, 6: 100. doi:10.3389/fenvs.2018.00100
- ^ Tabatabai A, Vallino JJ, Follows MJ (2018) "Application of maximum entropy production principle in the assessment biogeochemical models". Ocean Sciences Meeting, Oregon, 11–16 Feb 2018.
- ^ Sciubba, E. (2011) "What did Lotka really say? A critical reassessment of the 'maximum power principle'". Ecological Modelling, 222(8): 1347–1353. doi:10.1016/j.ecolmodel.2011.02.002
- ^ a b Kavanagh, L. and Galbraith, E. (2018) "Links between fish abundance and ocean biogeochemistry as recorded in marine sediments". PloS One, 13(8). doi:10.1371/journal.pone.0199420 A.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Andersen KH and Pedersen M (2010) "Damped trophic cascades driven by fishing in model marine ecosystems", Proceedings of the Royal Society of London B: Biological Sciences, 277(1682): 795–802. doi:10.1098/rspb.2009.1512.
- ^ Rathburn, A.E., Willingham, J., Ziebis, W., Burkett, A.M. and Corliss, B.H. (2018) "A New biological proxy for deep-sea paleo-oxygen: Pores of epifaunal benthic foraminifera". Scientific Reports, 8(1): 1–8. doi:10.1038/s41598-018-27793-4
- ^ Hutchins, D.A. and Fu, F. (2017) "Microorganisms and ocean global change". Nature microbiology, 2(6): 17058. doi:10.1038/nmicrobiol.2017.58.
- ^ *Ciais, P., Sabine, C., Bala, G., Bopp, L., Brovkin, V., Canadell, J., Chhabra, A., DeFries, R., Galloway, J., Heimann, M. and Jones, C. (2014) "Carbon and other biogeochemical cycles supplementary material". In: Climate change 2013: the physical science basis, pages 465–570, Cambridge University Press. ISBN 9781107057999.
- ^ Rhein M, Rintoul SR, Aoki S et al (2013) "Observations: Ocean". In: Stocker TF, Qin D, Plattner G-K et al (Eds.) Climate change 2013: the physical science basis, pages 255–316 Cambridge University Press. ISBN 9781107057999.
- ^ Käse, L. and Geuer, J.K. (2018) "Phytoplankton responses to marine climate change–an introduction". In YOUMARES 9-The Oceans: Our Research, Our Future, pages 55–71, Springer. doi:10.1007/978-3-319-93284-2_5.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Pimiento C, Leprieur F, Silvestro D, Lefcheck JS, Albouy C, Rasher DB, Davis1 M, Svenning JC and Griffin JN (2020) "Functional diversity of marine megafauna in the Anthropocene". Science Advances, 6(16): eaay7650. doi:10.1126/sciadv.aay7650
- ^ "Extinction of threatened marine megafauna would lead to huge loss in functional diversity." ScienceDaily, 17 April 2020.
- ^ Sievers, M., Brown, C.J., Tulloch, V.J., Pearson, R.M., Haig, J.A., Turschwell, M.P. and Connolly, R.M. (2019) "The role of vegetated coastal wetlands for marine megafauna conservation". Trends in ecology & evolution, 34(9). doi:10.1016/j.tree.2019.04.004.
The 100 µm length scale in the microbial ocean
[edit]
100 µm is a useful yardstick in the microscale world of marine bacteria. While environmental processes affecting microbial ecology often unfold at larger scales, as exemplified by the phytoplankton bloom depicted in blue hues in the background, it is the microscale that most immediately affects the life of marine and aquatic bacteria. Characteristic distances among bacteria, whether motile (flagellated cells) or non-motile (circular, non-flagellated cells), as well as between microbes and viruses occur over scales on the order of 100 µm, here denoted as ‘O(100 µm)’. So, too, do gradients of dissolved organic matter (yellow-green coloration and grey contour lines) emanating for example from phytoplankton (diatom on the top right), to which motile bacteria can respond by directional swimming (thick dashed trajectory). Processes such as turbulence (not depicted) also contribute to spatial heterogeneity at O(100 µm) scales, further increasing the prominence of this scale in microbial ecology.[1]]]
Turbulent stirring and molecular diffusion
[edit]"100 µm is a fundamental scale for chemical gradients that bacteria experience in the ocean even when sources are larger than 100 µm, because of turbulent stirring, which can rapidly create filaments and sheets of O(100 µm). Whereas molecular diffusion is unavoidable and its magnitude is dictated almost exclusively by the chemical in question, the presence and intensity of turbulence are site-dependent. Yet, in many regions of the ocean, turbulence is prevalent, and its effect in stirring chemicals is rather insensitive to its intensity. The most immediate action of turbulence — and a fundamental precursor to its long-term, intuitive effect of mixing — is to stir what is suspended in water. This stirring produces finer and finer concentration features, down to a smallest scale — the Batchelor scale — below which molecular diffusion takes over and obliterates chemical gradients into homogeneity. Stirring thus reshapes heterogeneities in the distribution of nutrients into a tangled web of nutrient sheets and filaments (Guasto et al. 2012, Taylor & Stocker 2012). The smallest and most prevalent sheets and filaments occur at the Batchelor scale, given by LB = (νD2/ε)1/4, where ν ~ 10−6 m2 s−1 is the kinematic viscosity of seawater and ε is the turbulent kinetic energy, a measure of the strength of turbulence. For many regions of the ocean, ε = 10−6 to 10−10 W kg−1. Therefore, for small molecules with D = 10−9 m2 s−1, LB = 30 to 300 µm, i.e. LB = O(100 µm)."[1]
Ecological interactions
[edit]
-
Microscopy and morphology
[edit]Traditional approach - Microscopy, cultivation and morphology
- Microscopy
- dark field microscopy
- bright field microscopy
- phase contrast microscopy
- polarized light microscopy
- electron microscopy
Metagenomics and DNA barcoding
[edit]Current approach
- Phylogenomic sampling
- Phylogenomic taxonomic sampling

Typically a tablespoon of seawater contains about 10 million viruses, one million bacteria, 10,000 algae, and 1,000 protists.[6]
These microorganisms are ubiquitous in the oceans, thriving at abyssal depths around superheated hydrothermal vents, flourishing in frozen sea ice floating at the surface of the Arctic Ocean, and abundant everywhere in between. "Altogether, all of the microbes in the sea (the majority of the plankton along with tiny crustaceans like copepods) outweigh the swimming animals (or nekton) like fish and whales by six to one".[6]
"Microbes in the ocean are extremely diverse, with thousands of new species discovered every year. This diversity allows these organisms to adapt to ever-changing environments, and likewise, the dynamic nature of the ocean enables the high diversity found there to flourish. Mostly, marine microbiologists use DNA sequencing rather than microscopy to identify microbes in the ocean. Many microbes that look the same under the microscope actually have very different DNA signatures, or barcodes. We are now excited to venture into the Gulf of Alaska and around the seamounts to see if we can discover new microbes there!"[6]
"In the Gulf of Alaska, we expect to find several thousand different microbes, but we only know what a few of them do! A 2002 NOAA Office of Ocean Exploration and Research expedition to the Gulf of Alaska Seamounts found numerous octocorals, which were found to be habitats for microbes (Penn et al. 2006, https://aem.asm.org/content/aem/72/2/1680.full.pdf (PDF, 167 KB). ) at depths of 600-3,300 meters (2,000 to 10,000 feet). Many of the bacteria found in that study were involved in sulfur cycling and some were previously identified associated with deep-sea sediments and hydrothermal vents"[6]
"The DNA sequencing techniques we use have come a long way since 2002, so instead of the 885 sequences that were recovered from the 2002 expedition, we expect to obtain at least 10,000 times more for the same price. This will allow us to gain a much better understanding of the microbiology of the Gulf of Alaska seamounts than has ever been possible before!"[6]
From Aquatic macroinvertebrate DNA barcoding: "DNA barcoding is an alternative method to the traditional morphological taxonomic classification... Since its introduction, the field of DNA barcoding has matured to bridge the gap between traditional taxonomy and molecular systematics. This technique has the ability to provide more detailed taxonomic information, particularly for cryptic, small, or rare species. DNA barcoding involves specific targeting of gene regions that are found and conserved in most animal species, but have high variation between members of different species."
From Molecular phylogenetics: "Molecular phylogenetics is one aspect of molecular systematics, a broader term that also includes the use of molecular data in taxonomy and biogeography... The most common approach is the comparison of homologous sequences for genes using sequence alignment techniques to identify similarity. Another application of molecular phylogeny is in DNA barcoding, wherein the species of an individual organism is identified using small sections of mitochondrial DNA or chloroplast DNA.
From: Taxonomy (biology): "The advent of molecular genetics and statistical methodology allowed the creation of the modern era of "phylogenetic systems" based on cladistics, rather than morphology alone."[7][8][9]
"DNA barcoding uses a short DNA sequence from a standardized and agreed-upon position in the genome as a molecular diagnostic for species-level identification."... Primarily barcodes "are very useful tools to accelerate species-level analysis of biodiversity and to facilitate conservation efforts. Barcodes have been used for identification of prey in gut contents, detection of invasive species, forensics, reveal cryptic species, and discover new species. Recent advances in sequencing technology allow the use of barcodes for rapid and increasingly automated biodiversity assessment."[10]
"Global changes are initiating a cascade of complex processes, which result, among other things, in global climate warming. Effects of global climate change are most pronounced in the Arctic, where the associate processes are progressing at a more rapid pace than in the rest of the world. Intensified transport of warmer water masses into the Arctic is causing shifts in species distributions and efforts to understand and track these change are currently intensified. However, Arctic marine fauna is the result of different recurring colonization events by Atlantic and Pacific Ocean populations, producing a very confounding evolutionary signal and making species identification by traditional morphological taxonomic analysis extremely challenging. In addition, many marine species are too small or too similar to identify reliably, even with profound taxonomic expertise. Nevertheless, the majority of current research focusing on artic marine communities still relies on the analysis of samples with traditional taxonomic methods, which tends to lack the necessary taxonomic, spatial and temporal resolution needed to understand the drastic ecosystem shifts underway. However, molecular methods are providing new opportunities to the field and their continuous development can accelerate and facilitate ecological research in the Arctic. Here, we discuss molecular methods currently available to study marine Arctic biodiversity, encouraging the DNA barcoding for improved descriptions, inventory and providing examples of DNA barcoding utilization in Arctic diversity research and investigations into ecosystem drivers."[11]
- Metagenomics
- Microbial DNA barcoding
- Algae DNA barcoding
- DNA barcoding in diet assessment
- DNA sequencing - gene sequencing
- references
- ^ a b Stocker, R. (2015) "The 100 µm length scale in the microbial ocean". Aquatic Microbial Ecology, 76(3): 189–194. doi:10.3354/ame01777.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Krabberød, A.K., Bjorbækmo, M.F., Shalchian-Tabrizi, K. and Logares, R. (2017) "Exploring the oceanic microeukaryotic interactome with metaomics approaches". Aquatic Microbial Ecology, 79(1): 1–12. doi:10.3354/ame01811.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Faust K and Raes J (2012) "Microbial interactions: From networks to models". Nat Rev Microbiol, 10: 538–550. doi:10.1038/nrmicro2832.
- ^ Kodzius, R. and Gojobori, T. (2015) "Marine metagenomics as a source for bioprospecting". Marine genomics, 24(1): 21–30. doi:10.1016/j.margen.2015.07.001.
- ^ Hug, Laura A.; Baker, Brett J.; Anantharaman, Karthik; Brown, Christopher T.; Probst, Alexander J.; Castelle, Cindy J.; Butterfield, Cristina N.; Hernsdorf, Alex W.; Amano, Yuki; Ise, Kotaro; Suzuki, Yohey; Dudek, Natasha; Relman, David A.; Finstad, Kari M.; Amundson, Ronald; Thomas, Brian C.; Banfield, Jillian F. (11 April 2016). "A new view of the tree of life". Nature Microbiology. 1 (5): 16048. doi:10.1038/nmicrobiol.2016.48. PMID 27572647. S2CID 3833474.
- ^ a b c d e Collins, R. Eric (2019) The Small and Mighty – Microbial Life NOAA: Ocean Exploration and Research, Gulf of Alaska Seamounts 2019 Expedition.
 This article incorporates text from this source, which is in the public domain.
This article incorporates text from this source, which is in the public domain.
- ^ Datta, Subhash Chandra (1988). Systematic Botany (4 ed.). New Delhi: New Age International. ISBN 978-81-224-0013-7. Retrieved 25 January 2015.
- ^ Stace, Clive A. (1989) [1980]. Plant taxonomy and biosystematics (2nd. ed.). Cambridge: Cambridge University Press. ISBN 978-0-521-42785-2. Retrieved 19 April 2015.
- ^ Stuessy, Tod F. (2009). Plant Taxonomy: The Systematic Evaluation of Comparative Data. Columbia University Press. ISBN 978-0-231-14712-5. Retrieved 6 February 2014.
- ^ The MarBOL Collection PLOS. Accessed 20 February 2020.
- ^ Walczyńska, K.S., Mańko, M.K. and Weydmann, A. (2018) "Arctic Ocean biodiversity and DNA barcoding a climate change perspective". In: YOUMARES 8–Oceans Across Boundaries: Learning from each other, pages 145–153, Springer. doi:10.1007/978-3-319-93284-2_10.
- further references
- Trivedi S, Rehman H Ansari AA, Saggu S, Panneerselvam C, Abbas ZH, Ahmad I, Ansari AA and Ghosh SK (2016) "DNA Barcoding in the Marine Habitat: An Overview". In: Trivedi S, Rehman H Ansari AA, Ghosh SK and Rehman H (Eds.) DNA Barcoding in Marine Perspectives: Assessment and Conservation of Biodiversity, pages 3–28, Springer. ISBN 9783319418407.
- Trivedi S, Ansari AA, Ghosh SK and Rehman H (2016) DNA Barcoding in Marine Perspectives: Assessment and Conservation of Biodiversity, Springer. ISBN 9783319418407.
Omics
[edit]- Omics
- Brüwer, J.D. and Buck-Wiese, H. (2018) Reading the Book of Life–Omics as a Universal Tool Across Disciplines. In: Jungblut S., Liebich V., Bode M. (Eds) YOUMARES 8–Oceans Across Boundaries: Learning from each other (pp. 73–82). Springer, Cham.
Nutricline
[edit]| Aquatic layers |
|---|
| Stratification |
| See also |
The nutricline is the cline or layer in an ocean or lake where the greatest change in the nutrient concentration is occurring with depth.[1]
ABSTRACT: "Global ocean primary production (PP) is a function of both light and nutrient availability. The vertical distribution of nutrients in the euphotic zone differs in both time and space. As a result, the vertical distribution of PP varies as well. Differences in the vertical distribution of PP have not, however, been systematically studied. Here, we focus on the open ocean and use nutricline depth, DNO3 (defined as [NO-3]=1μmol kg-1), as a proxy for nutrient availability in the euphotic zone. Using our own and archived (WOD, HOT, BATS, CARIACO) data, we show universal relationships between DNO3 and (1) depth of the deep chlorophyll maximum (DCM), (2) total water column PP, (3) vertical distribution of PP. When DNO3 is located between ~20 and 90 m, the DCM and DNO3 are juxtaposed. However, the DCM is located above nutriclines found at > ~90 m. The observed relationships between DCM and DNO3 depths can be explained with a simple model including light and nutrient limitation. The global primary production estimates indicate that ~25% of ocean PP occurs in the upper 10 m. Estimating total global ocean PP from surface optical characteristics and the relationship between vertical PP distribution and DNO3 indicates that oligotrophic regions of the ocean may be more productive than usually assumed. The relationship shown here between water column PP and DNO3 suggests that considering stratification characteristics in a future ocean is critical for predicting climate change effects on global PP."[9]
FROM: deep chlorophyll maximum...
The deep chlorophyll maximum (DCM), also called the subsurface chlorophyll maximum,[2][3] is the region below the surface of water with the maximum concentration of chlorophyll. A DCM is not always present - sometimes there is more chlorophyll at the surface than at any greater depth - but it is a common feature of most aquatic ecosystems, especially in regions of strong thermal stratification.[4] The depth, thickness, intensity, composition, and persistence of DCMs vary widely.[3][5] The DCM generally exists at the same depth as the nutricline.[1]


FROM: Ocean deoxygenation...
Ocean deoxygenation poses implications for ocean productivity, nutrient cycling, carbon cycling, and marine habitats.[8] Most of the excess heat from CO2 and other greenhouse gas emissions is absorbed by the oceans.[9] Warmer oceans cause deoxygenation both because oxygen is less soluble in warmer water,[10] and through temperature driven stratification of the ocean which inhibits the production of oxygen from photosynthesis.[11] The ocean surface stratifies as the atmosphere and ocean warms causing ice melt and glacial runoff. This results in a less salty and therefore a less dense layer that floats on top.[12] Also the warmer waters themselves are less dense. This stratification inhibits the upwelling of nutrients (the ocean constantly recycles its nutrients) into the upper layer of the ocean.[13] This is where the majority of oceanic photosynthesis (such as by phytoplankton) occurs.[14] This decrease in nutrient supply is likely to decrease rates of photosynthesis in the surface ocean, which is responsible for approximately half of the oxygen produced globally.[14] Increased stratification can also decrease the supply of oxygen to the interior of the ocean.
"Nutricline depth controls the coccolithophorid-to-diatom (C/D) ratio."[15]
FROM: coccolithophore...
Deep-dwelling coccolithophore species abundance is greatly affected by nutricline and thermocline depths. These coccolithophores increase in abundance when the nutricline and thermocline are deep and decrease when they are shallow.[16]
- See also
- References
- ^ a b Estrada, M; Marrasé, C; Latasa, M; Berdalet, E; Delgado, M; Riera, T (1993). "Variability of deep chlorophyll maximum characteristics in the Northwestern Mediterranean". Marine Ecology Progress Series. 92: 289–300. Bibcode:1993MEPS...92..289E. doi:10.3354/meps092289. ISSN 0171-8630.
- ^ Jochem, Frank J.; Pollehne, Falk; Zeitzschel, Bernt (January 1993). "Productivity regime and phytoplankton size structure in the Arabian Sea" (PDF). Deep Sea Research Part II: Topical Studies in Oceanography. 40 (3): 711–735. Bibcode:1993DSRII..40..711J. doi:10.1016/0967-0645(93)90054-q. ISSN 0967-0645.
- ^ a b Anderson, G. C. (May 1969). "Subsurface Chlorophyll Maximum in the Northeast Pacific Ocean1". Limnology and Oceanography. 14 (3): 386–391. Bibcode:1969LimOc..14..386A. doi:10.4319/lo.1969.14.3.0386. ISSN 0024-3590.
- ^ Weston, K.; Fernand, L.; Mills, D. K.; Delahunty, R.; Brown, J. (2005-09-01). "Primary production in the deep chlorophyll maximum of the central North Sea". Journal of Plankton Research. 27 (9): 909–922. doi:10.1093/plankt/fbi064. ISSN 1464-3774.
- ^ Cullen, JJ. (1982). "The Deep Chlorophyll Maximum: Comparing Vertical Profiles of Chlorophyll a". Canadian Journal of Fisheries and Aquatic Sciences. 39 (5): 791–803. doi:10.1139/f82-108.
- ^ Richardson, K. and Bendtsen, J. (2019). "Vertical distribution of phytoplankton and primary production in relation to nutricline depth in the open ocean". Marine Ecology Progress Series, 620: 33–46. doi:10.3354/meps12960
- ^ Wirtz, K. and Smith, S.L. (2020). "Vertical migration by bulk phytoplankton sustains biodiversity and nutrient input to the surface ocean". Nature, Scientific Reports, 10(1): 1–12. doi:10.1038/s41598-020-57890-2
- ^ Harvey, Fiona (2019-12-07). "Oceans losing oxygen at unprecedented rate, experts warn". The Guardian. ISSN 0261-3077. Retrieved 2019-12-07.
- ^ Levitus, Sydney, et al. "Warming of the world ocean." Science 287.5461 (2000): 2225–2229
- ^ https://www.ysi.com/File%20Library/Documents/Technical%20Notes/DO-Oxygen-Solubility-Table.pdf
- ^ "Climate-driven trends in contemporary ocean productivity." Nature 444.7120 (2006): 752–755
- ^ Sigman, Daniel M., Samuel L. Jaccard, and Gerald H. Haug. "Polar ocean stratification in a cold climate." Nature 428.6978 (2004): 59–63
- ^ Arrigo, Kevin R., et al. "Phytoplankton community structure and the drawdown of nutrients and CO 2 in the Southern Ocean." Science 283.5400 (1999): 365–367. Behrenfeld, Michael J., et al. "Climate-driven trends in contemporary ocean productivity." Nature 444.7120 (2006): 752–755
- ^ a b Cermeño, Pedro, et al. "The role of nutricline depth in regulating the ocean carbon cycle." Proceedings of the National Academy of Sciences 105.51 (2008): 20344-20349
- ^ Cermeno, P.; Dutkiewicz, S.; Harris, R. P.; Follows, M.; Schofield, O.; Falkowski, P. G. (2008). "The role of nutricline depth in regulating the ocean carbon cycle". Proceedings of the National Academy of Sciences. 105 (51): 20344–9. Bibcode:2008PNAS..10520344C. doi:10.1073/pnas.0811302106. JSTOR 25465827. PMC 2603260. PMID 19075222.
- ^ Kinkel, H.; et al. (2000), "Coccolithophores in the equatorial Atlantic Ocean: response to seasonal and Late Quaternary surface water variability", Marine Micropaleontology, 39 (1–4): 87–112, Bibcode:2000MarMP..39...87K, doi:10.1016/s0377-8398(00)00016-5
Marine food web
[edit]

Ecological pyramids are graphical representations, along the lines of the diagram at the right, which show how biomass or productivity changes at each trophic level in an ecosystem. The first or bottom level is occupied by primary producers or autotrophs (Greek autos = self and trophe = food). These are the names given to organisms that do not feed on other organisms, but produce biomass from inorganic compounds, mostly by a process of photosynthesis.
- Comparison of marine and terrestrial food webs
"Ecologists have greatly advanced our understanding of the processes that regulate trophic structure and dynamics in ecosystems. However, the causes of systematic variation among ecosystems remain controversial and poorly elucidated. Contrasts between aquatic and terrestrial ecosystems in particular have inspired much speculation, but only recent empirical quantification. Here, we review evidence for systematic differences in energy flow and biomass partitioning between producers and herbivores, detritus and decomposers, and higher trophic levels. The magnitudes of different trophic pathways vary considerably, with less herbivory, more decomposers and more detrital accumulation on land. Aquatic–terrestrial differences are consistent across the global range of primary productivity, indicating that structural contrasts between the two systems are preserved despite large variation in energy input. We argue that variable selective forces drive differences in plant allocation patterns in aquatic and terrestrial environments that propagate upward to shape food webs. The small size and lack of structural tissues in phytoplankton mean that aquatic primary producers achieve faster growth rates and are more nutritious to heterotrophs than their terrestrial counterparts. Plankton food webs are also strongly size-structured, while size and trophic position are less strongly correlated in most terrestrial (and many benthic) habitats. The available data indicate that contrasts between aquatic and terrestrial food webs are driven primarily by the growth rate, size and nutritional quality of autotrophs. Differences in food-web architecture (food chain length, the prevalence of omnivory, specialization or anti-predator defences) may arise as a consequence of systematic variation in the character of the producer community".[1]
- (2018) Comparison of marine and terrestrial ecosystems: suggestions of an evolutionary perspective influenced by environmental variation
- (2018) Why are marine ecosystems biologically more diversified than their equivalent terrestrial ecosystems?
- (2005) All wet or dried up? Real differences between aquatic and terrestrial food webs
- (2000) Are there real differences among aquatic and terrestrial food webs?
- (2012) Food Web: Concept and Applications
- (1996) Linking marine and terrestrial food webs: allochthonous input from the ocean supports high secondary productivity on small islands and coastal land communities
- Cited by 780
- "Terrestrial productivity varies from 3 tp 3,5000 g.m-2.yr-1 dry mass... In several areas, tremendously productive coastal waters are juxtaposed with unproductive land habitats. This contract is particularly evident in areas of great upwelling along arid coastlines, such as the Benguela current ecosystem and the hyper-arid Namib Dessert of western southern Arica..."
- Food Web National Geographic. <============ U S E
- "A food web consists of all the food chains in a single ecosystem. Each living thing in an ecosystem is part of multiple food chains. Each food chain is one possible path that energy and nutrients may take as they move through the ecosystem. All of the interconnected and overlapping food chains in an ecosystem make up a food web."
- Marine Food Chain National Geographic, 2010
- Trites, A.W. (2003) "Food webs in the ocean: who eats whom and how much". Responsible fisheries in the marine ecosystem, pp. 125–141. FAO. [10]
- Carrion cycling in food webs: comparisons among terrestrial and marine ecosystems
- (2003) Comparing marine and terrestrial ecosystems: implications for the design of coastal marine reserves pdf
- Cited by 445
- Grazers & Predators MarineBio Conservation Society. Last update: February 16, 2019
- Similarities, differences and mechanisms of climate impact on terrestrial vs. marine ecosystems Review
- Biomass Pyramid for a Marine Ecosystem
- Food chains and food webs
- Introducing Marine Ecosystems
- What is a Trophic Cascade?
From: Photosynthetic efficiency...
- Algae and other unicellular organisms: From a 2010 study by the University of Maryland, photosynthesizing Cyanobacteria have been shown to be a significant species in the global carbon cycle, accounting for 20–30% of Earth's photosynthetic productivity and convert solar energy into biomass-stored chemical energy at the rate of ~450 TW.[2] The efficiency of algae is much higher compared to that of plants (98 percent efficiency for algae compared to just 12 percent in plants).[3]
- Spring bloom – seasonal plankton blooms

Microorganisms
[edit]
Right side: microbial loop, with bacteria using dissolved organic carbon to gain biomass, which then re-enters the classic carbon flow through protists.[5]

-
Pelagibacter ubique, the most abundant bacteria in the ocean, plays a major role in the global carbon cycle


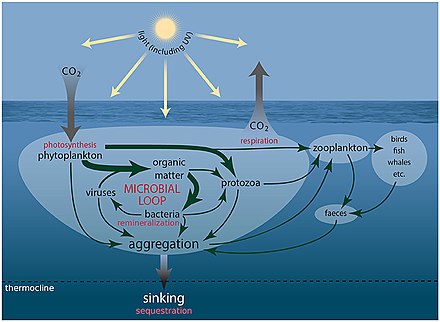
- Marine methane paradox
- New Study Explains Mysterious Source of Greenhouse Gas Methane in the Ocean Woods Hole Oceanographic Institution, 21 November 2016.
- Repeta, Daniel J. et al.(2016) "Marine methane paradox explained by bacterial degradation of dissolved organic matter". Nature Geoscience. doi:10.1038/ngeo2837.
Mixotrophs
[edit]Polar webs
[edit]
Definitions:[11]
- A food web is a group of predators and prey that interact in a habitat or ecosystem
- A stressor is a factor that reduces the health or productivity of an ecosystem (i.e., causes stress)
Antarctic
[edit]| Differences between the Arctic and the Antarctic regions | ||
|---|---|---|
| Arctic | Antarctic | |
| Geology | An ocean, partly frozen in the form of a massive ice cap. The Arctic ice cap floats on the surface of the Arctic Ocean, which plunges an average depth below of 1,000 metres.[10] | A frozen continent covered almost entirely with glaciers. The Antarctic has ice mountains that rise to 4,900 metres,[11] and is surrounded by the Southern Ocean. |
| Ocean currents and temperature | affected by warm waters of the gulf stream. A lot warmer than the Antarctic. | the ocean surrounding the Antarctic landmass is dominated by the Antarctic Circumpolar Current which allows little circulation of heat from other regions. This leads to extreme low temperatures (average -55 °C) and is the reason for the existing ice sheet. |
| Animal life | The arctic fox as well as marine mammals like polar bears, walrus, seals, and whales. Birds like puffins, fulmars, and other bird species. | Penguins and marine mammals like whales and seals |
| Human populations | Indigenous population (Inuits, Indians, Siberians) and enduring settlements | Historically deserted though today there are impermanent settlements for scientists. |

"Phaeocystis pouchetii... is one of the few non-diatom algae which is exceedingly plentiful in southern Antarctic waters. Phaeocystis occurs in either a unicellular motile form or a colonial palmelloid stage which can be large enough to be visible to the naked eye. This alga is very widely distributed in warm and temperate waters as well as in Antarctic and Arctic seas. Its gelatinous blooms are sometimes known as "Dutchman's baccy juice", and the jelly-like colonies are so thick that they clog plankton nets. Extensive blooms of Phaeocystis have been reported in the Ross Sea (El-Sayed et al., 1983) where high cell concentrations extended from the surface to depths of 100–150 m. Distinct discoloration of the water (greenish-grey to greenish-brown) accompanied this bloom, resulting in considerable submarine light attenuation."[15]
- FOR IMAGE --> Bender, S.J., Moran, D.M., McIlvin, M.R., Zheng, H., McCrow, J.P., Badger, J., DiTullio, G.R., Allen, A.E. and Saito, M.A. (2018) "Colony formation in Phaeocystis antarctica: connecting molecular mechanisms with iron biogeochemistry". Biogeosciences, 15(16). doi:10.5194/bg-15-4923-2018. full text
- The marine environment - Discovering Antarctica <======= E X P L O R E
- Antarctica Food Chains and Food Webs
Anthropogenic effects
[edit]From Limacina helicina... Pteropods are strict pelagic mollusks that are highly adapted to life in the open ocean.[16] They are actively swimming in the water. Limacina helicina is a holoplanktonic species. Habitat of Limacina helicina is upper epipelagic and glacial.[17] It lives in temperatures from -0.4 °C to +4.0 °C or rarely up to 7 °C.[18] Limacina helicina of the size from 0.2 to 0.4 mm lives mainly in depths from 0 m to 50 m.[19] Larger pteropods lives from 0 m to 150 m.[19] Already Constantine John Phipps mentioned its "innumerable quantities" in arctic seas in 1774.[20] Limacina helicina is a major component of the polar zooplankton.[21] It can comprise more than 50% of total zooplankton abundance (number of individuals per unit volume).[21] Species of the clade Thecosomata produce a fragile external calcium carbonate shell, which could serve as a [[Sailing The aragonitic composition of the shell makes it very sensitive to dissolution.[16] Aragonite is a metastable form of calcium carbonate and it is more soluble in seawater than calcite.[22] Because of its highly soluble[16] aragonite shell and polar distribution, Limacina helicina may be one of the first organisms affected by ocean acidification, and it is therefore a key indicator species of this process.[21] A decline of pteropod populations would likely cause dramatic changes to various pelagic ecosystems.[22][16] Shelled pteropods also play a geochemical role in carbon cycle in the oceans, as they contribute to the export of calcium carbonate and can represent a major component of the carbon transport to the deep ocean.[22][16][21] Researchers found 24-53% individuals of Limacina helicina with shells damaged by dissolution off the U.S. West Coast in 2011.[23]
See also
[edit]References
[edit]- ^ Shurin, J.B., Gruner, D.S. and Hillebrand, H. (2006) "All wet or dried up? Real differences between aquatic and terrestrial food webs". Proceedings of the Royal Society B: Biological Sciences, 273(1582): 1-9. doi:10.1098/rspb.2005.3377.
- ^ Pisciotta JM, Zou Y, Baskakov IV (2010). "Light-Dependent Electrogenic Activity of Cyanobacteria". PLOS ONE. 5 (5): e10821. doi:10.1371/journal.pone.0010821. PMC 2876029. PMID 20520829.
- ^ Ingenious ‘control panel’ in algae provides blueprint for super-efficient future solar cells
- ^ Phytoplankton NASA Earth Observatory. Accessed 30 November 2019.]
- ^ Krabberød, A.K., Bjorbækmo, M.F., Shalchian-Tabrizi, K. and Logares, R. (2017) "Exploring the oceanic microeukaryotic interactome with metaomics approaches". Aquatic Microbial Ecology, 79(1): 1–12. doi:10.3354/ame01811.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ DeLong E and Karl D (2005) "Genomic perspectives in microbial oceanography". Nature, 437: 336–342. doi:10.1038/nature04157.
- ^ Mateus, M.D. (2017) "Bridging the gap between knowing and modeling viruses in marine systems—An upcoming frontier". Frontiers in Marine Science, 3: 284. doi:10.3389/fmars.2016.00284.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ^ Benway, H.M., Lorenzoni, L., White, A.E., Fiedler, B., Levine, N.M., Nicholson, D.P., DeGrandpre, M.D., Sosik, H.M., Church, M.J., O’brien, T.D. and Leinen, M. (2019) "Ocean time series observations of changing marine ecosystems: An era of integration, synthesis, and societal applications". Frontiers in Marine Science, 6(393). doi:10.3389/fmars.2019.00393.
- ^ Siegel, D.A., Buesseler, K.O., Behrenfeld, M.J., Benitez-Nelson, C.R., Boss, E., Brzezinski, M.A., Burd, A., Carlson, C.A., D'Asaro, E.A., Doney, S.C. and Perry, M.J. (2016) "Prediction of the export and fate of global ocean net primary production: The EXPORTS science plan". Frontiers in Marine Science, 3: 22. doi:10.3389/fmars.2016.00022.
- ^ "The Mariana Trench – Oceanography". www.marianatrench.com. 4 April 2003. Archived from the original on 7 December 2006. Retrieved 2 December 2006.
- ^ Antarctica. In The Kingfisher Children's Encyclopedia. New York, New York: Kingfisher. 2012. p. 16.
- ^ Deppeler, S.L. and Davidson, A.T. (2017). "Southern Ocean Phytoplankton in a Changing Climate". Review, Frontiers in Marine Science, 4: 40. doi:10.3389/fmars.2017.00040.
- ^ Sullivan, C. W., McClain, C. R., Comiso, J. C., and Smith, W. O. (1988). "Phytoplankton standing crops within an Antarctic ice edge assessed by satellite remote sensing". J. Geophys. Res. 93:12487. doi:10.1029/JC093iC10p12487.
- ^ Petrou, K., Kranz, S. A., Trimborn, S., Hassler, C. S., Ameijeiras, S. B., Sackett, O., et al. (2016). "Southern Ocean phytoplankton physiology in a changing climate". J. Plant Physiol. 203: 135–150. doi:10.1016/j.jplph.2016.05.004.
- ^ - Antarctic Phytoplankton
- ^ a b c d e Comeau, S.; Jeffree, R.; Teyssié, J. L.; Gattuso, J. P. (2010). Stepanova, Anna (ed.). "Response of the Arctic Pteropod Limacina helicina to Projected Future Environmental Conditions". PLOS ONE. 5 (6): e11362. doi:10.1371/journal.pone.0011362. PMC 2894046. PMID 20613868.
- ^ Bouchet, P.; Gofas, S. (2011). Limacina helicina (Phipps, 1774). Accessed through: World Register of Marine Species at http://www.marinespecies.org/aphia.php?p=taxdetails&id=140223 on 2011-01-29
- ^ "Limacina helicina helicina helicina". Marine Species Identification Portal, accessed 8 February 2011.
- ^ a b Kobayashi, H. A. (1974). "Growth cycle and related vertical distribution of the thecosomatous pteropod Spiratella (?Limacina?) helicina in the central Arctic Ocean". Marine Biology. 26 (4): 295–301. doi:10.1007/BF00391513. S2CID 84668586.
- ^ Phipps C. J. (1774). A Voyage towards the North Pole undertaken by His Majesty's Command 1773. J. Nourse, London, viii + 253 pp. Page 195.
- ^ a b c d Hunt, B.; Strugnell, J.; Bednarsek, N.; Linse, K.; Nelson, R. J.; Pakhomov, E.; Seibel, B.; Steinke, D.; Würzberg, L. (2010). Finkel, Zoe (ed.). "Poles Apart: The "Bipolar" Pteropod Species Limacina helicina is Genetically Distinct Between the Arctic and Antarctic Oceans". PLOS ONE. 5 (3): e9835. doi:10.1371/journal.pone.0009835. PMC 2847597. PMID 20360985.
- ^ a b c Comeau, S.; Gorsky, G.; Jeffree, R.; Teyssié, J. -L.; Gattuso, J. -P. (2009). "Impact of ocean acidification on a key Arctic pelagic mollusc (Limacina helicina)". Biogeosciences. 6 (9): 1877–1882. doi:10.5194/bg-6-1877-2009.
- ^ Bednaršek, N.; Feely, R. A.; Reum, J. C. P.; Peterson, B.; Menkel, J.; Alin, S. R.; Hales, B. (2014). "Limacina helicina shell dissolution as an indicator of declining habitat suitability owing to ocean acidification in the California Current Ecosystem". Proceedings of the Royal Society B. 281 (1785): 1785. doi:10.1098/rspb.2014.0123. PMC 4024287. PMID 24789895.
External links
[edit]- Ocean Color Time Series NASA
- Upwelling (Italian wikipedia)
Mixotrophic plankton
[edit]Mixotrophic plankton that combine phototrophy and heterotrophy – table based on Stoecker et. al., 2017 [1]
| |||||||
|---|---|---|---|---|---|---|---|
| General types | Description | Example | Further examples | ||||
| Bacterioplankton | Photoheterotrophic bacterioplankton | 
|
Vibrio cholerae | Roseobacter spp. Erythrobacter spp. Gammaproteobacterial clade OM60 Widespread among bacteria and archaea | |||
| Phytoplankton | Called constitutive mixotrophs by Mitra et. al., 2016.[2] Phytoplankton that eat: photosynthetic protists with inherited plastids and the capacity to ingest prey. | 
|
Ochromonas species | Ochromonas spp. Prymnesium parvum Dinoflagellate examples: Fragilidium subglobosum,Heterocapsa triquetra,Karlodinium veneficum,Neoceratium furca,Prorocentrum minimum | |||
| Zooplankton | Called nonconstitutive mixotrophs by Mitra et. al., 2016.[2] Zooplankton that are photosynthetic: microzooplankton or metazoan zooplankton that acquire phototrophy through chloroplast retentiona or maintenance of algal endosymbionts. | ||||||
| Generalists | Protists that retain chloroplasts and rarely other organelles from many algal taxa | 
|
Most oligotrich ciliates that retain plastidsa | ||||
| Specialists | 1. Protists that retain chloroplasts and sometimes other organelles from one algal species or very closely related algal species | 
|
Dinophysis acuminata | Dinophysis spp. Myrionecta rubra | |||
| 2. Protists or zooplankton with algal endosymbionts of only one algal species or very closely related algal species | 
|
Noctiluca scintillans | Metazooplankton with algal endosymbionts Most mixotrophic Rhizaria (Acantharea, Polycystinea, and Foraminifera) Green Noctiluca scintillans | ||||
- ^ Stoecker, D.K., Hansen, P.J., Caron, D.A. and Mitra, A. (2017) "Mixotrophy in the marine plankton". Annual Review of Marine Science, 9: 311–335. doi:10.1146/annurev-marine-010816-060617
- ^ a b Mitra A, Flynn KJ, Tillmann U, Raven J, Caron D, et al (2016) "Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition; incorporation of diverse mixotrophic strategies". Protist, 167: 106–20. doi:10.1016/j.protis.2016.01.003
Locomotion of protists
[edit]
Protists are often categorised according to their mode of locomotion. Most unicellular protists are motile and can generate movement using flagella, cilia or pseudopods. Cells which use flagella are usually referred to as flagellates, cells which use cilia for movement are usually referred to as ciliates, and cells which use pseudopods are usually referred to as amoeba or amoeboids.
Flagellates include bacteria as well as protists. The rotary motor model used by bacteria uses the protons of an electrochemical gradient in order to move their flagella. Torque in the flagella of bacteria is created by particles that conduct protons around the base of the flagellum. The direction of rotation of the flagella in bacteria comes from the occupancy of the proton channels along the perimeter of the flagellar motor.[1]
Ciliates use small flagella called cilia to power their movement through the water. One ciliate will generally have hundreds to thousands of cilia that are densely packed together in arrays. During movement, an individual cilium deforms using a high-friction power stroke followed by a low-friction recovery stroke. Since there are multiple cilia packed together on an individual organism, they display collective behavior in a metachronal rhythm. This means the deformation of one cilium is in phase with the deformation of its neighbor, causing deformation waves that propagate along the surface of the organism. These propagating waves of cilia are what allow the organism to use the cilia in a coordinated manner to move. A typical example of a ciliated microorganism is the Paramecium, a one-celled, ciliated protozoan covered by thousands of cilia. The cilia beating together allow the Paramecium to propel through the water at speeds of 500 micrometers per second.[2]
Protists according to how they move
| |||||||
|---|---|---|---|---|---|---|---|
| Type of protist | Movement mechanism | Description | Example | Other examples | |||
| Flagellate | use one or several flagella. A flagella is a lash-like appendage that protrudes from the cell body of some protists (as well as some bacteria). | 
|
Cryptophytes | Excavates are considered to be the most basal flagellate lineage.[3] Other flagellates include dinoflagellates, choanoflagellates, most green algae, some radiolarians (probably gametes), foraminiferans (as gametes) | |||
| Ciliate | use multiple cilia, which are small flagella, to power themselves through the water | 
|
Paramecium bursaria - click to expand and see cillia | Foraminiferans, and some marine amoebae, ciliates and flagellates. | |||
| Amoeba (Amoeboids) |
Amoeba have the ability to alter shape by extending and retracting pseudopods.[4] | 
|
Amoeba proteus | are found in every major lineage of protists. Amoeboid cells occur not only among the protozoa, but also in fungi, algae, and animals.[5][6] | |||
Haeckel, Turing and radiolarian morphology
[edit]closely replicate some radiolarian shell patterns [7]
Radiolarians are unicellular protists with elaborate silica shells



- Kunstformen der Natur (Art Forms in Nature)
- The Chemical Basis of Morphogenesis
Turing, A.M. (1990) "The chemical basis of morphogenesis". Bulletin of mathematical biology, 52(1-2): 153–197. SEE: http://www.dna.caltech.edu/courses/cs191/paperscs191/turing.pdf
- Turing patterns on a sphere, 1999...
"Turing systems were originally put forward to explain the chemical basis of morphogenesis, that is, how a zygote, which is a spherical object, can acquire a form with a smaller symmetry. Our calculations are ideal to revive this discussion. Very simple unicellular organisms present complicated symmetry breaking that should be compared with Turing’s predictions. Among the simplest ones are the Radiolaria. These micro-organisms present very beautiful patterns of silicates formed on their membranes. There is an incredible variety of forms, some are spherical and others are conical or elongated. Many, although not all, of the radiolaria skeletons have hexagonal structuring on their surfaces. In Fig. 7 we show photographs of two selected common spherical radiolaria. Notice the resemblance of the skeleton in Fig. 7!a" with our pattern in Fig. 5, and the remarkable correspondence of the skeleton in Fig. 7!b" with our pattern in Fig. 6"...
". The skeleton of microscopic sea animals, called Radiolaria, have spherical form with motifs arranged in tetrahedral, octahedral, icosahedral, and other more complex symmetries #6$. Notably, the skeleton of a Radiolarian called Aulonia hexagona shows a fairly distorted triangular lattice spread out over the sphere !see Fig. 55 in Ref. #11$". This pattern contains also distorted pentagons as in the example we give in Fig. 6."
Numerical analyses have already been performed by Varea et al. [11], and their results compare favorably with the structures of various radiolaria.
computer matches were very good
Pattern formation on a sphere
skeletal patterns in radiolaria allows us to compare theoretical results with spherical biological systems such as the skeletons of several radiolaria
Their simulations show the emergence of different symmetries on this surface and are reminiscent of skeletal patterns in radiolaria
Radiolarian patterns can arise naturally from a uniform sphere
FROM: Patterns in nature... Ernst Haeckel (1834–1919) painted beautiful illustrations of marine organisms, in particular Radiolaria, emphasising their symmetry to support his faux-Darwinian theories of evolution....[9] In 1952, Alan Turing (1912–1954), better known for his work on computing and codebreaking, wrote The Chemical Basis of Morphogenesis, an analysis of the mechanisms that would be needed to create patterns in living organisms, in the process called morphogenesis.[10] He predicted oscillating chemical reactions, in particular the Belousov–Zhabotinsky reaction. These activator-inhibitor mechanisms can, Turing suggested, generate patterns (dubbed "Turing patterns") of stripes and spots in animals, and contribute to the spiral patterns seen in plant phyllotaxis.[11]
a seminal paper... which was subsequently shown to predict the type of patterns found in radiolarians, such as in the photo below.
- Bernard Richards
- [12]
- pioneering work on morphogenesis[13]
- The Turing Guide -
Turing’s theory of morphogenesis
FROM: https://www.sciencedaily.com/releases/2016/08/160809095259.htm "Geologists appreciate the radiolarian ooze very much, because the alien-looking creatures have lived on our planets for approximately 500 million years. Therefore, fossil radiolarians are very useful for dating geological structures... "Ernst Haeckel and other former natural scientists classified the radiolarians according to their morphology, that is their appearance in a broad sense. But it is not always true that organisms are genetically related even if they have a similar appearance," Krabberød explains... Already around 1870, Ernst Haeckel discovered a photosynthesizing algae living in colonies together with radiolarians, in a common gel secreted by the radiolarian hosts. Organisms living together like this are called symbionts. Anders Krabberød has, by using DNA analysing techniques, shown that several marine species in the protistan alveolate group(link is external) also seem to be living as symbionts with radiolarians, but these are hiding themselves inside the siliceous exoskeletons of certain species... Radiolarians are exclusive salt water species and can be found in every ocean on Earth. Some of them have pseudopodia -- "false feet" -- that can be used for different purposes."
FROM: https://blogs.scientificamerican.com/artful-amoeba/proteus-how-radiolarians-saved-ernst-haeckel/ "Radiolarians are tiny protists that live inside intricate silica shells. Because silica is impervious to the acids that often dissolve shells made of calcium carbonate at great depth, they make up a huge proportion of the sludge found on deep sea beds. They extend tiny pseudopods out from their shell to capture food, and sometimes they house algae to help feed them... They were among the earliest eukaryotes to evolve at the end of the Pre-Cambrian, 550 million years ago, and their shells have varied so much over time that they are useful for dating petroleum beds and geological formations. But why do they have those shells, and what force could be selecting for the endless variety in their structure? I do not know... "
FROM: https://www.sciencedirect.com/topics/earth-and-planetary-sciences/radiolaria "Radiolaria are single-celled marine planktonic protozoa that secrete an opal skeleton composed of a number of architectural elements (radial spicules, internal bars, external spines) that are joined together to form regular symmetrical structures. The skeletons are usually smaller than 2 mm and commonly between 100 and 250 μm in diameter. Radiolaria can be solitary and colonial, the latter producing centimetre- to metre-sized aggregates (Flügel, 2004). Radiolaria are found exclusively in marine sediments, proving some information about the nature of soil parent materials (Clarke, 2003; Stoops, 2003)."
translucent cage over 500 million years old, amongst the earliest skeletonized life forms
Calcite and silicate
[edit]From calcium carbonate...
"In warm, clear tropical waters corals are more abundant than towards the poles where the waters are cold. Calcium carbonate contributors, including plankton (such as coccoliths and planktic foraminifera), coralline algae, sponges, brachiopods, echinoderms, bryozoa and mollusks, are typically found in shallow water environments where sunlight and filterable food are more abundant. Cold-water carbonates do exist at higher latitudes but have a very slow growth rate. The calcification processes are changed by ocean acidification... Where the oceanic crust is subducted under a continental plate sediments will be carried down to warmer zones in the asthenosphere and lithosphere. Under these conditions calcium carbonate decomposes to produce carbon dioxide which, along with other gases, give rise to explosive volcanic eruptions... The carbonate compensation depth (CCD) is the point in the ocean where the rate of precipitation of calcium carbonate is balanced by the rate of dissolution due to the conditions present. Deep in the ocean, the temperature drops and pressure increases. Calcium carbonate is unusual in that its solubility increases with decreasing temperature. Increasing pressure also increases the solubility of calcium carbonate. The carbonate compensation depth can range from 4,000 to 6,000 meters below sea level."
"Sediment cores from the ocean floor contain types of information that scientists use to better understand the fluctuations of global climate. Perhaps the most important information is that gleaned from the microscopic shells of animals such as this, called planktonic Foraminifera ("forams" for short). Forams provide two main types of information. First of all, different species of forams prefer different ocean temperature and nutrient conditions. . Scientists can therefore learn much about the climatic conditions of a core site in the past by looking at which species once inhabited the area. Secondly, the shells of forams effectively lock in the oxygen and carbon isotopic composition of the waters in which they formed. Because past periods of glaciation changed the relative quantities of heavy oxygen (18O) and light oxygen (16O), scientists can use the isotopic composition of foram shells as a proxy signal for past changes in global ice volume. Other chemical measures are available as well by studying the composition of the shells. Data from ocean cores about past glaciation matches Milankovitch's theory remarkably well."
In many parts of the world, silica is the major constituent of sand.[14]
From silicon dioxide...
"For well over a billion years, silicification in and by cells has been common in the biological world. In the modern world it occurs in bacteria, single-celled organisms, plants, and animals (invertebrates and vertebrates). Prominent examples include:
- Tests or frustules (i.e. shells) of diatoms, Radiolaria and testate amoebae.[15]
- Silica phytoliths in the cells of many plants, including Equisetaceae, practically all grasses, and a wide range of dicotyledons.
- The spicules forming the skeleton of many sponges."
From: Biomineralization

"Biomineralization is the process by which living organisms produce minerals,[a][17] often to harden or stiffen existing tissues. Such tissues are called mineralized tissues. It is an extremely widespread phenomenon; all six taxonomic kingdoms contain members that are able to form minerals, and over 60 different minerals have been identified in organisms.[18][19][20] Examples include silicates in algae and diatoms, carbonates in invertebrates, and calcium phosphates and carbonates in vertebrates. These minerals often form structural features such as sea shells and the bone in mammals and birds. Organisms have been producing mineralised skeletons for the past 550 million years. Ca carbonates and Ca phosphates are usually crystalline, but silica organisms (sponges, diatoms...) are always non crystalline minerals. Other examples include copper, iron and gold deposits involving bacteria. Biologically-formed minerals often have special uses such as magnetic sensors in magnetotactic bacteria (Fe3O4), gravity sensing devices (CaCO3, CaSO4, BaSO4) and iron storage and mobilization (Fe2O3•H2O in the protein ferritin).
Biomineralization: Complete conversion of organic substances to inorganic derivatives by living organisms, especially micro-organisms.[21]
The first evidence of biomineralization dates to some 750 million years ago,[22][23] and sponge-grade organisms may have formed calcite skeletons 630 million years ago.[24] But in most lineages, biomineralization first occurred in the Cambrian or Ordovician periods.[25] Organisms used whichever form of calcium carbonate was more stable in the water column at the point in time when they became biomineralized,[26] and stuck with that form for the remainder of their biological history[27] (but see [28] for a more detailed analysis). The stability is dependent on the Ca/Mg ratio of seawater, which is thought to be controlled primarily by the rate of sea floor spreading, although atmospheric CO2 levels may also play a role.[26]"
| Type of mineralization | Examples of organisms |
|---|---|
| Calcium carbonate (calcite or aragonite) | |
| Silica |
|
| Apatite (phosphate carbonate) |
|
Images
[edit]





from coccolithophore, terrigenous sediment and pelagic sediment...
"Where biogenic constituents compose less than 30 percent of the total, the deposit is called a deep-sea clay, brown mud, or red clay".
The skeletons are elaborate external hard mineral shells, usually made of silica (as in radiolaria and diatoms) or of calcium carbonate (as in foraminifera and coccoliths).[29]
- radiolarians
Radiolarians are unicellular with diameters from 0.1 to 0.2 mm, and grow intricate mineral skeletons. They are found as zooplankton throughout the ocean, and their skeletal remains make up much of the cover of the ocean floor as siliceous ooze. Due to their rapid species turn-over, they are important diagnostic microfossils found from the Cambrian onwards.
- diatoms
- foraminiferans
FROM: https://ucmp.berkeley.edu/foram/foramintro.html "Foraminifera (forams for short) are single-celled protists with shells. Their shells are also referred to as tests because in some forms the protoplasm covers the exterior of the shell. ... Radiating from the opening are fine hairlike reticulopodia, which the foram uses to find and capture food... A single individual may have one or many nuclei within its cell. The largest living species have a symbiotic relationship with algae, which they "farm" inside their shells. Other species eat foods ranging from dissolved organic molecules, bacteria, diatoms and other single celled phytoplankton, to small animals such as copepods. They move and catch their food with a network of thin extensions of the cytoplasm called reticulopodia, similar to the pseudopodia of an amoeba, although much more numerous and thinner."
FROM: https://www.ncbi.nlm.nih.gov/pubmed/14601411 "Foraminiferal research lies at the border between geology and biology. Benthic foraminifera are a major component of marine communities, highly sensitive to environmental influences, and the most abundant benthic organisms preserved in the deep-sea fossil record. These characteristics make them important tools for reconstructing ancient oceans."
Most foraminiferans have calcareous tests, composed of calcium carbonate.[30] A few forams have tests made of organic material, and in one genus of silica. Tests as fossils are known from as far back as the Cambrian period,[31] and many marine sediments are composed primarily of them. For instance, the limestone that makes up the pyramids of Egypt is composed almost entirely of nummulitic benthic Foraminifera.[32] It is estimated reef Foraminifera generate about 43 million tons of calcium carbonate per year.[33]
foraminifera FROM: https://www.youtube.com/watch?v=JLSa8cGJixQ foraminifera go back about 650 years ago The typical foraminifera is about the size of a period at the end of a sentence... foraminifera reflect environmental change - they are very sensitive indicators of environmental change. One way foraminifera can tell us something is by chemical analysis of the shells. You can look at the isotopic composition of the oxygen and the carbon and trace element concentrations in the shell. That means we can say things about direct temperature of the past. It can tell us about the polar ice caps. It can tell something about how much photosynthesis was going on in the surface of the ocean, and that can tell us something about the CO2 levels in the atmosphere.
See Paleocene–Eocene Thermal Maximum for data derives from foraminifera fossils
foraminifera are big players in constructing what we know about palaeoclimatology.
Genetic studies have identified the naked amoeba Reticulomyxa and the peculiar xenophyophores as foraminiferans without tests.
Most foraminifera species are benthic, though 40 morphospecies are planktonic.[34]
Planktonic foraminifera are represented by many species with worldwide occurrence in broad latitudinal and temperature belts, floating in the surface or near-surface waters of the open ocean as part of the marine zooplankton. Benthic foraminifera are as successful as the planktonic foraminifera group and even more abundant in modern seas and can live attached or free, at all depths.
- coccolithophores
Ooze is pelagic sediment that consists of at least 30% of microscopic remains of either calcareous or siliceous planktonic debris organisms. Oozes can be defined by and classified according to the predominate organism that compose them. For example, there are diatom, coccolith, foraminifera, globigerina, pteropod, and radiolarian oozes. Oozes are also classified and named according to their mineralogy, i.e. calcareous or siliceous oozes. Whatever their composition, all oozes accumulate extremely slowly, at no more than a few centimeters per millennium.[35][36]
Calcareous ooze is ooze that is composed of at least 30% of the calcareous microscopic shells—also known as tests—of foraminifera, coccolithophores, and pteropods. This is the most common pelagic sediment by area, covering 48% of the world ocean's floor. This type of ooze accumulates on the ocean floor at depths above the carbonate compensation depth. It accumulates more rapidly than any other pelagic sediment type, with a rate that varies from 0.3–5 cm/1000 yr.[37][35]
Siliceous ooze is ooze that is composed of at least 30% of the siliceous microscopic "shells" of plankton, such as diatoms and radiolaria. Siliceous oozes often contain lesser proportions of either sponge spicules, silicoflagellates or both. This type of ooze accumulates on the ocean floor at depths below the carbonate compensation depth. Its distribution is also limited to areas with high biological productivity, such as the polar oceans, and upwelling zones near the equator. The least common type of sediment, it covers only 15% of the ocean floor. It accumulates at a slower rate than calcareous ooze: 0.2–1 cm/1000 yr.[37][35]
Diatoms are divided into two groups that are distinguished by the shape of the frustule: the centric diatoms and the pennate diatoms.
Pennate diatoms are bilaterally symmetric. Each one of their valves have openings that are slits along the raphes and their shells are typically elongated parallel to these raphes. They generate cell movement through cytoplasm that streams along the raphes, always moving along solid surfaces.
Centric diatoms are radially symmetric. They are composed of upper and lower valves - epitheca and hypotheca - each consisting of a valve and a girdle band that can easily slide underneath each other and expand to increase cell content over the diatoms progression. The cytoplasm of the centric diatom is located along the inner surface of the shell and provides a hollow lining around the large vacuole located in the center of the cell. This large, central vacuole is filled by a fluid known as "cell sap" which is similar to seawater but varies with specific ion content. The cytoplasmic layer is home to several organelles, like the chloroplasts and mitochondria. Before the centric diatom begins to expand, its nucleus is at the center of one of the valves and begins to move towards the center of the cytoplasmic layer before division is complete. Centric diatoms have a variety of shapes and sizes, depending on from which axis the shell extends, and if spines are present.
Misc
[edit]-
Simple Food Web
-
Central Baltic Food Web Model
-
build up of toxins in a food chain
-
Carbonate-Silicate Cycle (Carbon Cycle focus)
-
Carbon-Slicate Cycle Feedbacks
-
Silica cycle draft
-
Marine Silica Cycle
Algae
[edit]-
Flagellar pit of Pyramimonas sp.
-
Flagellar pit of Pyramimonas sp.
-
Two Cryptophytes
-
Cryptophytes under light microscope
-
Colony of Pediastrum algae (freshwater)
-
green algae Pteromonas
-
Green algae, Chlorophyceae - chlorophytes
-
Colony of green alga, Pediastrum (light micrograph) (freshwater)
-
Spirotaenia, a genus of basal green algae that may be sister to the Chlorokybophyceae
-
Haeckel Siphoneae
-
Green algae on coastal rocks
-
Photomicrograph of multiple strands of Stigeoclonium
-
Green algae?
-
Haeckel Melethallia
-
largest marine genera
-
Acetabularia acetabulum
-
Brown algae
-
kelp beds
-
Red algae, Cyanidium sp.
Penard Collection
[edit]-
Amoeba from the Amoeba Collection made by Eugène Penard at the beginning of the 20th century
- Pamphagus granulatus
-
A Rhizaria covered in diatoms, from Eugène Penard's Amoeba Collection made by at the beginning of the 20th century
misc protozoa
[edit]-
Stramenopile (heterokont) have two, unequal flagella
-
Fucus receptacle with sterile paraphysesz, oogonium, and an ostiole where the opening in the surface of the receptacle is
-
Lingulodinium polyedrum, a species of photosynthetic dinoflagellates that can cause red tides and bioluminescent displays on beaches
-
Electron microscope image of Chlamydomonas reinhardtii, a single-cell green alga about that swims with two flagella - used as a model organism for research on fundamental questions in cell and molecular biology
-
Sea lettuces are edible green algae common along the coasts
-
Chroococcidiopsis is one of the most primitive cyanobacteria known. It can photsynthesize in far-red light, and might be suitable for future colonising Mars.
-
Klebsormidium bilatum are charophyte green alga forming multicellular, non-branching filaments
Choanoflagellate
[edit]Choanoflagellates - a group of free-living unicellular and colonial flagellate eukaryotes considered to be the closest living relatives of the animals.
Marine diatoms
[edit]-
Inner and external plates of a single valve of the centric diatom Coscinodiscus wailesii
-
diatoms
-
diatom
-
Diatom, Helipelta metil
-
Diatom, Triceratium favus
-
diatoms
-
diatoms
-
diatoms
-
diatom
-
diatom
-
Centric diatom
-
diatoms
-
diatoms
-
centric diatom
-
diatoms
-
pennate diatom
-
pennate diatom
-
pennate diatom
-
various diatoms
-
diatoms
-
diatoms
-
diatoms ##
-
diatoms
-
diatoms
-
diatoms
-
Closterium sp. (with unknown diatom), mitotic phase
-
Biddulphia pulchella
-
Biddulphia sp.
-
The diatom Phaeodactylum tricornutum can grow in the absence of silicon, and can survive without making silicified frustules.
-
Diatom
-
The pennate diatom Fragilariopsis kerguelensis is the most abundant diatom in the Antarctic Seas".[38]
-
Diatom Isthmia nervosa
-
Haeckel Diatomea
-
Haeckel Diatomea
-
Diatom Odontella aurita
-
Pleurosigma angulatum diatoms
-
Diatom frustle
-
Диатомит Инзенского месторождения, электронный микроскоп
-
Diatom
-
Diatom - Isthmia nervosa
-
Diatom - Champylodiscus sp
Radiolarians
[edit]-
Radiolarian protists drawn by Haeckel 1904
-
Radiolarians (Haeckel)
-
Radiolarian
-
Fossil radiolarian
-
Challenger
-
Haeckel Radiolaria: Spumellaria
-
Haeckel Cyrtoidea
-
Haeckel Phaeodaria
-
Haeckel Acanthometra
-
Radiolaria - Sijthoff
-
Haeckel - Discoidea
-
Haeckel Phaeodaria
-
-
Radiolarians
Unidentified radiolarians
[edit]-
(not identified)
-
Halicalyptra petalospyris ?
Coccolithophores
[edit]Foraminiferans
[edit]-
Foraminiferan
-
Test of a planktic foraminiferan with defensive spines
-
Test of Globigerina bulloides, an ooze-forming protozoan foraminiferan that is found as marine plankton
-
Benthic foram, Elphidium incertum
-
Benthic foram, Elphidium excavatum clavatum
-
Haeckel Thalamophora
-
Haeckel Thalamophora
-
Haeckel Thalamophora
-
Planktonic foram
-
Calcareous Foraminifera
-
Elphidium, benthic foraminifera, about 400 µm ø
-
Planktonic foraminifera
-
Foraminifera
-
Foraminifera
-
Foraminifera
-
Foraminifera
-
Orbitolina texana foraminifera fossils
-
fossil foraminifera
-
Textilaria, a genus of benthic foraminifera
-
Live Ammonia tepida streaming granular ectoplasm for catching food etc
-
Dentalina commonis, which belongs to an order Lagenide of benthic foraminifera that have a finely perforated radial calcitic shell , in which the crystals appear wrapped by organic membranes.
-
section of Rotalia beccarii
-
Operculina section
Dinoflagellates
[edit]-
Karenia brevis
-
Dinoflagellates
-
The free-living unarmored dinoflagellata
-
The free-living unarmored dinoflagellata
-
The free-living unarmored dinoflagellata
-
Haeckel Peridinea
-
Colonial dinoflagellates
-
Planktonic dinoflagellate, Ornithocercus magnificus
-
Many dinoflagellates are encased in cellulose armor and have two flagella that fit in grooves between the plates. Movement of these two perpendicular flagella causes a spinning motion.
Ciliates and amoeba
[edit]-
Ciliate Ciliophora
-
Suctoria sucking another ciliate
-
Ciliate Ciliophora
-
Tintinnid Tintinnopsis campanula
-
Euplotes with amoeba
Euglena
[edit]-
Drawing of a Euglena single cell flagellate (fresh and saltwater)
Gastrotricha
[edit]-
Gastrotricha
-
Gastrotricha
-
Gastrotricha
Fungi
[edit]-
Haeckel - Mycetozoa (slime moulds)
Gastropods
[edit]-
Undescribed species of deepwater nudibranch
Miscellaneous
[edit]-
Bathyphysa conifera, sometimes called the "flying spaghetti monster", a bathypelagic species of siphonophore
-
Reflectivity from silvery fish
-
Microfossils
-
Protozoa
-
Planktonic jellyfish Medusae drift in the water column and move by contracting the muscles within the bell-like structure creating pulse-like movements. The tentacles contain harpoon-like stinging cells to capture small planktonic shrimp and crabs.
-
Ctenophore
-
Radiata diversity
-
Hydra (freshwater only)
-
Onychodictyon, extinct lobopodan
-
Diagram of a typical animal cell. Organelles are labelled as follows: (1) Nucleolus; (2) Nucleus; (3) Ribosome; (4) Vesicle; (5) Rough endoplasmic reticulum; (6) Golgi apparatus; (7) Microtubule; (8) Smooth endoplasmic reticulum; (9) Mitochondrion; (10) Vacuole; (11) Cytoplasm; (12) Lysosome; (13) Centrioles
-
Marine siphonophore
-
phytoplankton
-
Cyanobacteria Dolichospermum crassum
-
Cyanobacteria Snowella
-
Dinoflagellat (Neoceratium horridum) with diatom (Coscinodiscus radiatus)
- ^ Brady, Richard M. (1993). "Torque and switching in the bacterial flagellar motor. An electrostatic model". Biophysical Journal. 64 (4): 961–973. Bibcode:1993BpJ....64..961B. doi:10.1016/S0006-3495(93)81462-0. PMC 1262414. PMID 7684268.
- ^ Lauga, Eric; Thomas R Powers (25 August 2009). "The hydrodynamics of swimming microorganisms". Reports on Progress in Physics. 72 (9): 096601. doi:10.1088/0034-4885/72/9/096601. S2CID 3932471.
- ^ Dawson, Scott C; Paredez, Alexander R (2013). "Alternative cytoskeletal landscapes: cytoskeletal novelty and evolution in basal excavate protists". Current Opinion in Cell Biology. 25 (1): 134–141. doi:10.1016/j.ceb.2012.11.005. PMC 4927265. PMID 23312067.
- ^ Singleton, Paul (2006). Dictionary of Microbiology and Molecular Biology, 3rd Edition, revised. Chichester, UK: John Wiley & Sons. p. 32. ISBN 978-0-470-03545-0.
- ^ David J. Patterson. "Amoebae: Protists Which Move and Feed Using Pseudopodia". Tree of Life web project.
- ^ "The Amoebae". The University of Edinburgh. Archived from the original on 10 June 2009.
- ^ Varea, C., Aragon, J.L. and Barrio, R.A. (1999) "Turing patterns on a sphere". Physical Review E, 60(4): 4588. doi:10.1103/PhysRevE.60.4588
- ^ Varea, C., Aragon, J.L. and Barrio, R.A.(1999) "Turing patterns on a sphere". Physical Review E, 60(4): 4588. doi:10.1103/PhysRevE.60.4588
- ^ Ball 2009, p. 41.
- ^ Turing, A. M. (1952). "The Chemical Basis of Morphogenesis". Philosophical Transactions of the Royal Society B. 237 (641): 37–72. Bibcode:1952RSPTB.237...37T. doi:10.1098/rstb.1952.0012. S2CID 120437796.
- ^ Ball 2009, pp. 163, 247–250.
- ^ Richards, Bernard (2005). "Turing, Richards and morphogenesis". The Rutherford Journal. 1.
- ^ Richards, Bernard (2017). "Chapter 35 – Radiolaria: Validating the Turing theory". In Copeland, Jack; et al. (eds.). [[The Turing Guide]]. pp. 383–388. ISBN 978-0-19-874782-6.
{{cite book}}: URL–wikilink conflict (help) - ^ Berslien E (2012). An Introduction to Forensic Geoscience. Wiley & Sons. p. 138. ISBN 9781405160544.
- ^ Cite error: The named reference
Fernwas invoked but never defined (see the help page). - ^ Vert, Michel; Doi, Yoshiharu; Hellwich, Karl-Heinz; Hess, Michael; Hodge, Philip; Kubisa, Przemyslaw; Rinaudo, Marguerite; Schué, François (2012). "Terminology for biorelated polymers and applications (IUPAC Recommendations 2012)" (PDF). Pure and Applied Chemistry. 84 (2): 377–410. doi:10.1351/PAC-REC-10-12-04. S2CID 98107080.
- ^ Harris, Ph.D., Edward D. (1 January 2014). Minerals in Food Nutrition, Metabolism, Bioactivity (1st ed.). Lancaster, PA: DEStech Publications, Inc. p. 378. ISBN 978-1-932078-97-8. Retrieved 30 January 2015.
- ^ Astrid Sigel; Helmut Sigel; Rol, K.O. Sigel, eds. (2008). Biomineralization: From Nature to Application. Metal Ions in Life Sciences. Vol. 4. Wiley. ISBN 978-0-470-03525-2.
- ^ Weiner, Stephen; Lowenstam, Heinz A. (1989). On biomineralization. Oxford [Oxfordshire]: Oxford University Press. ISBN 978-0-19-504977-0.
- ^ Jean-Pierre Cuif; Yannicke Dauphin; James E. Sorauf (2011). Biominerals and fossils through time. Cambridge. ISBN 978-0-521-87473-1.
- ^ Vert, Michel; Doi, Yoshiharu; Hellwich, Karl-Heinz; Hess, Michael; Hodge, Philip; Kubisa, Przemyslaw; Rinaudo, Marguerite; Schué, François (11 January 2012). "Terminology for biorelated polymers and applications (IUPAC Recommendations 2012)". Pure and Applied Chemistry. 84 (2): 377–410. doi:10.1351/PAC-REC-10-12-04. S2CID 98107080.
- ^ Porter, S. (2011). "The rise of predators". Geology. 39 (6): 607–608. Bibcode:2011Geo....39..607P. doi:10.1130/focus062011.1.
- ^ Cohen, P. A.; Schopf, J. W.; Butterfield, N. J.; Kudryavtsev, A. B.; MacDonald, F. A. (2011). "Phosphate biomineralization in mid-Neoproterozoic protists". Geology. 39 (6): 539–542. Bibcode:2011Geo....39..539C. doi:10.1130/G31833.1.
- ^ Maloof, A. C.; Rose, C. V.; Beach, R.; Samuels, B. M.; Calmet, C. C.; Erwin, D. H.; Poirier, G. R.; Yao, N.; Simons, F. J. (2010). "Possible animal-body fossils in pre-Marinoan limestones from South Australia". Nature Geoscience. 3 (9): 653–659. Bibcode:2010NatGe...3..653M. doi:10.1038/ngeo934.
- ^ Wood, R. A. (2002). "Proterozoic Modular Biomineralized Metazoan from the Nama Group, Namibia". Science. 296 (5577): 2383–2386. Bibcode:2002Sci...296.2383W. doi:10.1126/science.1071599. ISSN 0036-8075. PMID 12089440. S2CID 9515357.
- ^ a b Zhuravlev, A. Y.; Wood, R. A. (2008). "Eve of biomineralization: Controls on skeletal mineralogy" (PDF). Geology. 36 (12): 923. Bibcode:2008Geo....36..923Z. doi:10.1130/G25094A.1.
- ^ Porter, S. M. (Jun 2007). "Seawater chemistry and early carbonate biomineralization". Science. 316 (5829): 1302. Bibcode:2007Sci...316.1302P. doi:10.1126/science.1137284. ISSN 0036-8075. PMID 17540895. S2CID 27418253.
- ^ Maloof, A. C.; Porter, S. M.; Moore, J. L.; Dudas, F. O.; Bowring, S. A.; Higgins, J. A.; Fike, D. A.; Eddy, M. P. (2010). "The earliest Cambrian record of animals and ocean geochemical change". Geological Society of America Bulletin. 122 (11–12): 1731–1774. Bibcode:2010GSAB..122.1731M. doi:10.1130/B30346.1.
- ^ Grobe, H., Diekmann, B. and Hillenbrand, C.D. (2009) "The memory of the Polar Oceans". In: Hempel, G. (Ed) Biology of Polar Oceans.
- ^ Cite error: The named reference
Sen_Gupta83was invoked but never defined (see the help page). - ^ Dartnell L (8 May 2008). "Sea creatures had a thing for bling". New Scientist. No. 2655.
- ^ Foraminifera: History of Study, University College London, retrieved 20 September 2007
- ^ Langer, M. R.; Silk, M. T. B.; Lipps, J. H. (1997). "Global ocean carbonate and carbon dioxide production: The role of reef Foraminifera". Journal of Foraminiferal Research. 27 (4): 271–277. doi:10.2113/gsjfr.27.4.271.
- ^ Hemleben, C.; Anderson, O.R.; Spindler, M. (1989). Modern Planktonic Foraminifera. Springer-Verlag. ISBN 978-3-540-96815-3.
- ^ a b c HüNeke, H., and T. Mulder (2011) Deep-Sea Sediments. Developments in Sedimentology, vol. 63. Elsiever, New York. 849 pp. ISBN 978-0-444-53000-4
- ^ Neuendorf, K.K.E., J.P. Mehl, Jr., and J.A. Jackson, J.A., eds. (2005) Glossary of Geology (5th ed.). Alexandria, Virginia, American Geological Institute. 779 pp. ISBN 0-922152-76-4
- ^ a b Rothwell, R.G., (2005) Deep Ocean Pelagic Oozes, Vol. 5. of Selley, Richard C., L. Robin McCocks, and Ian R. Plimer, Encyclopedia of Geology, Oxford: Elsevier Limited. ISBN 0-12-636380-3
- ^ Hart, T.J (1942). "Phytoplankton periodicity in Antarctic surface waters". Discovery Rep.
Cite error: There are <ref group=lower-alpha> tags or {{efn}} templates on this page, but the references will not show without a {{reflist|group=lower-alpha}} template or {{notelist}} template (see the help page).

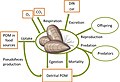

























![Mesodinium rubrum produce deep red blooms using enslaved chloroplasts from their algal prey [5]](http://upload.wikimedia.org/wikipedia/commons/thumb/d/df/Mesodinium_rubrum.jpg/270px-Mesodinium_rubrum.jpg)






![Opal can contain protist microfossils of diatoms, radiolarians, silicoflagellates and ebridians [15]](http://upload.wikimedia.org/wikipedia/commons/thumb/d/d9/Coober_Pedy_Opal_Doublet.jpg/352px-Coober_Pedy_Opal_Doublet.jpg)
![Marble can contain protist microfossils of foraminiferans, coccolithophores, calcareous nannoplankton and algae, ostracodes, pteropods, calpionellids and bryozoa [15]](http://upload.wikimedia.org/wikipedia/commons/thumb/6/61/MarmoCipollino_FustoBasMassenzioRoma.jpg/320px-MarmoCipollino_FustoBasMassenzioRoma.jpg)




![Cosmarium, a genus of freshwater green algae from which the land plants emerged.[6][7]](http://upload.wikimedia.org/wikipedia/commons/thumb/3/37/Cosmarium201512081550.JPG/226px-Cosmarium201512081550.JPG)








![Venus flytrap photosynthesing and eating insects[30]](http://upload.wikimedia.org/wikipedia/commons/thumb/2/2f/Venus_flytrap_plant_photosynthesing_and_eating_insects.png/291px-Venus_flytrap_plant_photosynthesing_and_eating_insects.png)
![Mixotrophic phytoplankton Karlodinium capturing and ingesting a small cell[30]](http://upload.wikimedia.org/wikipedia/commons/thumb/a/a1/Mixotrophic_phytoplankton_Karlodinium_capturing_and_ingesting_a_small_cell.png/271px-Mixotrophic_phytoplankton_Karlodinium_capturing_and_ingesting_a_small_cell.png)
![The mixotroph Dinophysis feeding on Mesodinium. The small red circles inside the Mesodinium are algae the Mesodinium has already eaten. The arrow points to the small straw or feeding tube it uses to feed on Mesodinium.[30]](http://upload.wikimedia.org/wikipedia/commons/thumb/e/e0/Mixotroph_Dinophysis_feeding_on_Mesodinium.png/313px-Mixotroph_Dinophysis_feeding_on_Mesodinium.png)

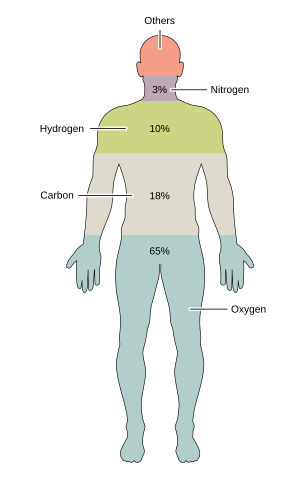



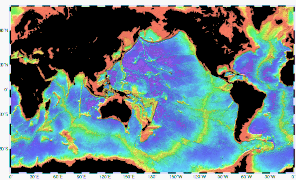
![Salinity [2]](http://upload.wikimedia.org/wikipedia/commons/thumb/6/61/SeaSurfaceSalinity.jpg/288px-SeaSurfaceSalinity.jpg)

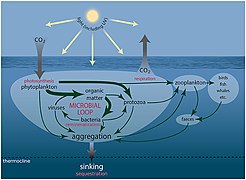












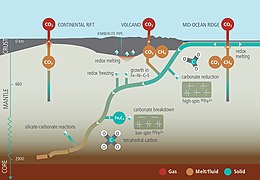

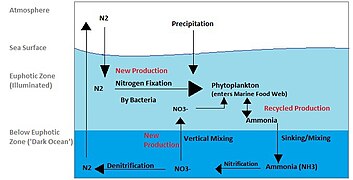


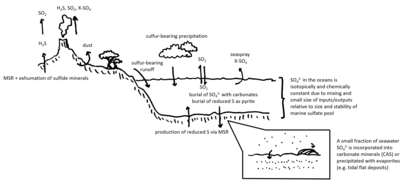

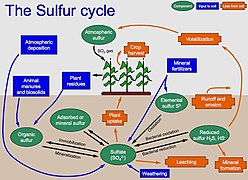



![Water cycle [3]](http://upload.wikimedia.org/wikipedia/commons/thumb/b/bc/Cycle_342.jpg/227px-Cycle_342.jpg)



















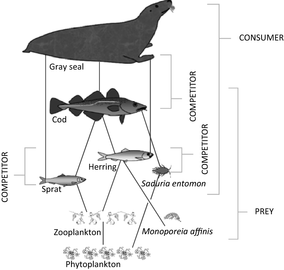

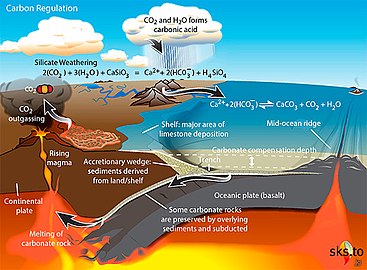

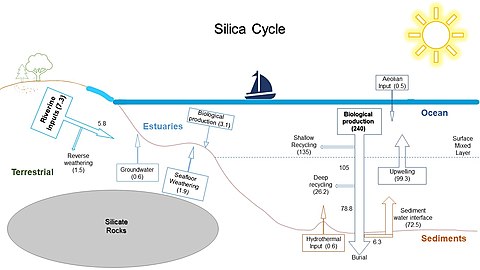
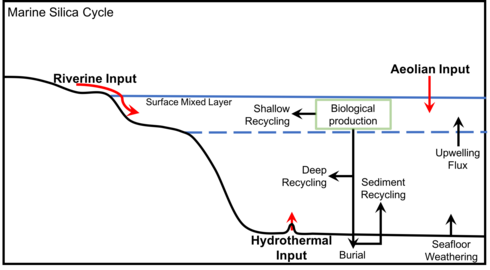

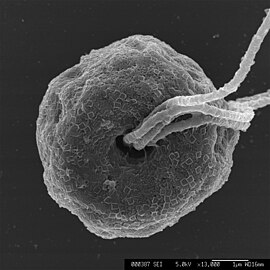


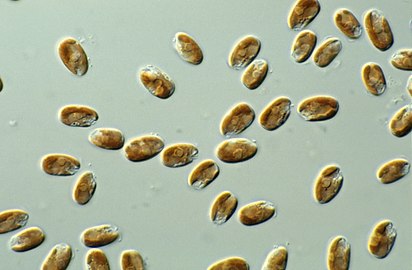







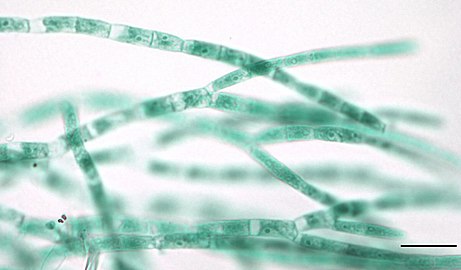





































































































![The pennate diatom Fragilariopsis kerguelensis is the most abundant diatom in the Antarctic Seas".[38]](http://upload.wikimedia.org/wikipedia/commons/thumb/1/16/Nitzschia-kerguelensis_hg.jpg/327px-Nitzschia-kerguelensis_hg.jpg)