Talk:Human genome/Archive 3
| This is an archive of past discussions about Human genome. Do not edit the contents of this page. If you wish to start a new discussion or revive an old one, please do so on the current talk page. |
| Archive 1 | Archive 2 | Archive 3 |
Concept → limitations
One possible way to resolve the above controversy over the "concept of the human genome" is to instead focus on the limitations on how we define the human genome and the practical consequences of those limitations. While Ken Weiss in The Scientist opinion piece did write about the genome as a "Platonic ideal", he also illustrated how the genome deviates from that ideal with specific examples. The advantage of focusing examples and limitations is that it is (1) closer to the sources, (2) it will be easier to understand by a general audience, and (3) is more relevant to the main topic of this article.
From Agricolae's analysis above, which I agree with, the following sentence is noncontroversial:
- Some find the phrase of "the human genome" represents a Platonic ideal, arguing that "'the human genome' that we have labeled as such doesn't actually exist".
- Source: Weiss K (2012-08-17). "What Is the Human Genome?". Opinion. The Scientist.
To which we could add:
Extended content
|
|---|
|
Thoughts? Boghog (talk) 08:54, 25 July 2013 (UTC)
- Boghog: Thanks for considering my ideas further. As for me, I am done with editing on this page. The most antagonistic departmental discussions are preferable to the way dialogue tends (not) to occur on Wikipedia. However, I will make one very tiny note, given your extensive and valuable commentary: Ken Weiss did not illustrate how "the genome" deviates from that ideal. He said that genomes deviate from the ideal. While I started out by fixating, then continued to fixate, on the odd obsession on this page with the monolithic generic use of "the", a linguistic construction which favors absolute objectivist truths, it is a broader point--that using simplifications obscures good science and creates bad science-that I am largely concerned with. Nobody has been able to defend the value of the phrase "the human genome" beyond the value of simplicity. My goal wasn't a grand debate about abstractions or even normality, but some editors dig their teeth into *the oddest thing*. With more suggestions like yours above, perhaps the article editors will collectively reach a point I would be happy with, even in my absence. I'm sure some clarification of the kind you provide would be helpful to non-technical readers, who might be currently wondering, if there is only one human genome, how can everyone have it and be different from it. Best wishes and good luck. Dylan Hunt (talk) 18:55, 25 July 2013 (UTC)
==Supplementary talk of genetic filaments---the physical function test and findings == 1, To bring back life with health by their direct relatives saw this and felt sad and cried.
Around 2-3 yrs ago, I read in a newspaper in US, a patient with cardiac problem was declared death by the hospital, but he came back to life afterwards. There are around 22-23% cardiac patients coming back to life after the death declaration, and this phenomenon is yet not well explained.
My explanation for this: when these patients were resuscitated, their direct relatives saw this and felt sad and cried. These sad cries came into the patients’ brains and resonated their double spiral filaments of the DNA. The same thing happened in the cardiac cells as well. And this led to the resurgence of the patients. During the resuscitation, the electric defibrillator must have been used, but this is only a single extrinsic factor. If we compare the defibrillator with the genetic filament resonance; first of all, they differ in being the intrinsic and extrinsic factors, second, the cries of the relative should provide the same frequency as the patients’ genetic filaments, and so the filaments are easier to resonate. Thus the resonance would have better effect and make resurgence easier. We declare death at the stop of the respiration and heart beat, but when the body temperature is still higher than 35 degree, the cells in the heart an brain are still alive, so the genetic filaments can still sense and resonate with the microwave from the external world, and the resurgence is still possible. But it requires their own children as it requires the same frequency and amplitude of the wave. If they are wife, daughter-in-law, son-in-law or brothers and sisters, the frequency and amplitude would vary and the resonance would be less effective, and the resurgence rate would only be 25% in those who can be resurged.
In June 2013 in yahoo news, “three professors in Oxford University were frozen to wait for resurgence”. Two of them had their heads cut and the third one was frozen as a whole.
In my opinion, the first two cannot be resurged while the third holds some possibilities, but it should be validated by multiple experiments with the mouse. Of course, if they were already dead due to severe disease, that would be another story.
Four to 5 decades ago when I was living in the north, I saw goldfish frozen in the glass bottle. After 1 day or two, when the ice melted, the goldfish could come back to life again. I also picked up one frozen fish in the lake in a winter, and when I put it back to water at home, it came back to life and swam again. The low cold blood animals could be resurged in this way, but if we cut their head when they were frozen, would they come back to life and swim again?
Human are higher animals, when they are sick, before they were really dead in all cells, if we finish all the process of preparation of resurgence, and when later they need to be resurged, it requires the blood circulation back gain, thus the cells could function and the genetic filaments would open and resonate with the wave exactly of the same frequency and amplitude from the external world. The hormone effect should be added as well. However, the electric field varies from one person to another, so the start and maintenance of the resonance would be a challenging task which needs at least 30 years of effort, lets wait and see.
2, The association of genetic filaments in plants with the external world;
A hundred year ago, we learned heredity secret lying in DNA from the bean experiment, but we never elaborated the close association between the DNA and the genetic filaments interaction.
In the Epoch Times on Jan 11th 2013, there was a piece of news titled “plants are like humans”, also in the newspaper of Kanzhongguo (www.kanzhonguo.com) in June 28th 2013, there was a paper titled “Thirsty! The first time that human heard the ‘scream’ of trees.”
In “plants are like humans”, it mentioned the extraordinary experiment done by lie detective Baxter in the sixties of the last century, that the plants could sense, and could have emotions as humans detected by lie detectors.
The happiness: when we watered the tongue orchid, the detector sensed a decrease wave as in humans
The terror: when we twitched the leaf and put it into hot coffee, it didn't change obviously, but if we tried to burn the leaf with the fire, even just with the thinking in mind would make a great shake in the wave and it surged to the peak. So we could know “my god, it knows what we are thinking!”
The super-sense: (here is extracted from the original article)“Baxter carried out an experiment like this: he threw several shrimps into hot water alive, and the plants were immediately irritated. The same response was recorded after several times. To exclude the man-made interference and to confirm the accuracy of the experiment, he used a newly designed machine and threw the shrimp into hot water in random time, and used a very delicate recorder to record the result. Baxter locked the 3 plants in 3 rooms and connected them with electrode, and no one was allowed to enter the rooms.”
The next day, when he went in to check the result, he found the waves were soaring 6-7 seconds after every throw of the shrimps into the water. All the 3 plants showed response, so Baxter said; we could almost affirm that plants have communications among them. In Yale University, Baxter put a spider and a plant into the same room, and the spider climbed onto the plant. But the recorder recorded a miracle--- before the spider came onto the plant, the plant had already showed some response. Obviously, the plant could super-sense the intention of the spider. The ones who ever doubted the experiments became the supporters: the experiment of Baxter surprised the world. The Doctor of Chemistry Mike in California thought the result was ridiculous. In order to challenge the result, he also did some experiments, but then his attitude changed absolutely. Mike also demonstrated that plant could sense human’s mind, which is to say, the plant can think as well, and feel the emotions of humans. So the plant is like human? Then where are their eyes? Why they could communicate their feelings? why they would feel the happiness, the terror or even super-sense? The experiments proved that they do have emotions, independently, and emotions don’t exist only in human or animals. The ancient Chinese people also always believed that “everything has spirit”, and they had fairy tales of the god of flower, the god of tree, telling that they have spirit like humans. In the article of “Thirsty! The first time that human heard the ‘scream’ of trees”: Just as human being are breathing desperately for air in dry season, the trees in draught, they would take as much the humidity as they could and give out the sound of “pah pah”. This sound is of a frequency 100 times more than the higher limit of frequency that human being could perceive. In my opinion: bean, tongue orchid and the trees are all plants, they have their own genes, so they would definitely have their DNA and double spiral filaments. All the connection between higher or lower mammals, birds, insects, fish, turtles, and plants, are based on resonance between double spiral filaments. They exchange information via it: happiness, sadness or terrors, or even the approach to death. All the species, as long as they have genes, they could send out and receive information with the double spiral filaments. The experiment of Baxter done 50 years ago, has supplemented my article of “genotype” published in Wikipedia between Dec 2009 and 2012. It proved the physical function of the double spiral filaments---they exchange information by waves, it can go across the wall and communicate between plants, it can go across oceans, go across half of the planet, it can go between the animals, or the animals with plants. The basic knowledge explained in the article in 2009 will not be explained here again. (Talker: Q. Y. Zhang; 香港 張其澐; --119.237.48.70 (talk) 00:27, 9 October 2013 (UTC))
Detail analysis of double spiral filaments in DNA beside function
One inspiration came over to my mind in one morning. Why DNA would contain two flat spiral filaments besides the 4 types of bases and thus form a “spring”in this way? And why they are hinging interlocked with the base groups, what do they serve for? And why there are also line style interlocks between C-G and A-T? I have designed great many machines including variant forms of the hinging spirals spring, so I am very familiar with the interlock theory. The dual long spirals spring could resonate in 3 dimensions, so the resonance could pass on in all directions. With the hinging interlock between base groups, the resonance could be achieved in the narrow intercellular space without affecting the functions of their own. Generally, even only with the cells along the sulci in the temporal lobes, the resonance could be started. It would be enough if each cell could open 4-5 base groups. ( possibility there are 3-5 Nos. cells death.) Not only for animals, they could also send out the microwave telepathy between human beings. The trumpet shape of the cerebral sulci enhances the force of sending and receiving the resonance, which further enables reception and communication between species and genus. ( Talker : Q. Y. Zhang; 香港 張其澐;--96.237.238.212 (talk) 21:04, 10 February 2014 (UTC))
==Natural selection---“random” and “non-random”
The biological field always pays great interest to new species emerged from “natural selection”. But when it comes to the analysis of gender rules, the human intelligence reaction, or the intelligence difference between siblings, they only conclude roughly with the word of “random”.
For example, if we ask a agriculturist, whether we would get a female or male baby in a pregnant horse or cow (the animal which always gives only one child at a time), he would say as the book tells “it’s random”. If we ask a doctor of the baby gender in a pregnant woman, he would say “It’s random” as well, (of course, they would not know the love and pursue process between this couple, and how this couple thought and reacted to the environment during the pregnancy). “Random” is the applied-to-all word to everything before we could find any rules our of it.
In “origins of species”, Darwin admitted that the species are always evolving; generation after generation, and he also advocated that the main propelling power the evolvement is the “natural selection”. The natural selection means that, when an individual gains some minimal physiological changes, and if thesis changes could benefit it more than its parents in survival and reproduction, this change would pass on and makes the following generations always on the direction of “better adapting to the environment and survival competition”. This theory includes: the gender of the species, the balance of the sex, and the principle of selecting a partner, and some trivial adjustment. Also it includes the balance and supplement to the hetero-gender, intelligence, characteristics and appearance.
The “rule of balance” persists for thousands of years of evolution. We now discuss only the “random and non-random” things in the biological filed.
The plants, including flowers, grass and trees, if they grow up, they need the sunshine, the rain and the soil. This is the rule of plant growing----the animals, they grow strong, they react fast and they run quickly, these are also the rules of “natural selection”. They could survive, and follow the rule of the biological food chain.The human beings could survive for thousands of years, there must also be some reproductive rules.
The animals and human beings would not go into distinction due to the single gender; this is the effect of reproductive rule. I have been studying on this for over 30 years, and finally make out a reasonable hypothesis. The hypothesis includes the rule of gender, and also how you could get a reactive and intelligent child with genetic advantages since fetus.* And the “random” rule in general ideas for determining the intelligence among siblings, now becomes a “non-random” concept
Before we could dig deeper for some rules, “random” is always a good word to explain all.But our species exist on this earth for thousands of years, and there must be many rules regulating them. Let us work together to better find out these thousands of rules.
- To see: www.gwencorp.com.hk/index.
(Talker: Q. Y. Zhang; 香港 張其澐--173.76.134.54 (talk) 16:48, 6 March 2014 (UTC))
Human, ape genome difference
Its not 4% its 1%. Please correct mistake. "Ever since researchers sequenced the chimp genome in 2005, they have known that humans share about 99% of our DNA with chimpanzees, making them our closest living relatives."
http://news.sciencemag.org/plants-animals/2012/06/bonobos-join-chimps-closest-human-relatives --109.23.159.201 (talk) 06:23, 21 May 2014 (UTC)
- The above source appears to be in error. It appears to refer to earlier outdated estimates "which were made using shorter alignable sequence fragments" (see PMID 16339373). Boghog (talk) 06:50, 21 May 2014 (UTC)
- Most of the articles talk about 1-2% of difference.
http://www.livescience.com/20940-unraveling-bonobo-genome-secrets.html http://en.wikipedia.org/wiki/Bonobo--109.23.159.201 (talk) 08:05, 21 May 2014 (UTC)
It depends on how the difference was measured. Many of the analyses are restricted to protein coding genes where the conservation is much higher (on the order of 99%). The variations in sequence in between genes (the so called junk DNA) is much lower (on the order of 95%). "Non-coding" DNA has a large influence on when and how genes are expressed, hence these differences are important to include in any between genome comparisons. The following analysis is particularly relevant:
- Larry Moran. "What's the Difference Between a Human and Chimpanzee?". Sandwalk.
This value of 1.5%, rounded up to 2%, gave rise to the widely quoted statement that humans and chimps are 98% identical. Britton (2002) challenged that number by pointing out that humans and chimp genomes differed by a large number of insertions and deletions (indels) that could not have been detected in hybridization studies. He claimed that there was an addition 3.4% of the genome that differed due to indels. That means the the real difference between humans and chimps is closer to 5% and we are only 95% identical!
{{cite web}}: Cite has empty unknown parameter:|1=(help); External link in|quote=
β-globin
The β-globin is on chromosome 11 and I think β-globin is too short a description.--Mark v1.0 (talk) 20:46, 5 November 2014 (UTC)
- Hi Mark v1.0. I'm afraid I don't understand where you're coming from. You're referring to this edit, in which you added "on chromosome 11" to the cell containing "β-globin" in the table at Human_genome#Human_genetic_disorders. The title of that column is "Chromosome or gene involved". Other cells in that column contain the name of a single gene, eg "APC", "Huntingtin", "CFTR". Do you think those cells should also include what chromosome the gene is on? Or do you think β-globin should be presented differently? Adrian J. Hunter(talk•contribs) 10:30, 7 November 2014 (UTC)
- Human_β-globin_locus says it is five genes, so you can not call it a single gene. What is your problem with having (chromosome 11) next to it?--Mark v1.0 (talk) 14:58, 9 November 2014 (UTC)
50% banana???
First, the banana genome is only about 16% the size of the human genome (523 million bp vs 3.2 billion bp). “Bananas (Musa spp.), including dessert and cooking types, are giant perennial monocotyledonous herbs of the order Zingiberales, a sister group to the well-studied Poales, which include cereals … Here we describe the draft sequence of the 523-megabase genome of a Musa acuminata doubled-haploid genotype, providing a crucial stepping-stone for genetic improvement of banana. “ (The banana (Musa acuminata) genome and the evolution of monocotyledonous plants, Angélique D’Hont, et al., Nature 488,213–217(09 August 2012)
Second, when I looked, I found the original claim, which has become ‘mutated’ over the years, was that bananas share 50% of our genes. In humans, there is a huge difference between sharing 50% of our genes and sharing 50% of our DNA, since less than 2% of the human DNA comprises genes (that is, less than 2% codes for protein).
Third, the original claim was made before either species’ genome had been sequenced.
What I found was that one scientist made an off-the-cuff rough guesstimate about how many genes bananas and humans share, and that somehow became ‘well-known scientific fact’. And not just that, but it became 'mutated' into 'well-known scientific fact' about how much DNA humans and bananas share.
To the best of my knowledge, the claim that humans and bananas share 50% of our DNA is little more than an urban legend or sorts, and does not belong on the page unless a valid scientific source can be found for it. — Preceding unsigned comment added by 2601:442:C180:3E13:E094:2427:C161:7BFE (talk) 11:02, 26 December 2015 (UTC)
- Thanks for the nice sleuthwork, anon. This is why we shouldn't source vague and outlandish claims to crummy British tabloids. I've removed the statement. Adrian J. Hunter(talk•contribs) 13:05, 26 December 2015 (UTC)
Edit Request
Such an important and topical subject and we get rubbish discussing the dead coming back to life on the talk page. This talk page seems to have been vandalized, since it starts out discussing something which seems quite relevant to why I went to the Human genome wikipedia article in the first place. It was well known by the scientific field that "the" Human genome as it was known in 2001 was nowhere near "complete". The claims and implications of this article are that we do know it, or just as bad, we know it "for all practical purposes". Give me a break; we have NO idea what practical purposes may result from discoveries in the "rest" of the genome (the 'rest' being the parts yet to be determined). The article fails miserably to explain what DNA is for the general reader. It is often considered just nuclear DNA (non-mt), but I agree that including mt-DNA is appropriate. What isn't appropriate is the clear implication that there is some unique human genome. I don't understand why this simplistic approach was used here. This stupidity spills over into discussion of chromosomes; we all do NOT have 22 pairs and one X, and half of us have another X and half of us have a Y sex chromosome. I'm not even well informed about genetic mutations, but I know enough to say that this is wrong; there are plenty of humans walking around with more (or less?) than this, not to mention the existence of mozaics, etc. I came to this article with the following questions: 1. What is the (known) range of size of the human genome in terms of base pairs? 2. How much of the ~1.5% of protein coding DNA consists of copies of one of the 'base set' of genes? 3.What percent of our genes are estimated have been inserted from other (non-primate, ie bacterial or viral) lineages? 4. What percent of those are believed to be under positive selection pressure (ie "useful")? Its a shame that I can find none of this here.Abitslow (talk) 18:48, 23 March 2016 (UTC)
An image for multiple gene-related pages
One thing on which few info can be found is on how the chromosomes connect to the DNA. There are images here, for example on the chromosome page, but these show an X-like structure. Same goes for other images to be found online:
- http://dnatestingexpert.com/wp-content/uploads/2013/09/DNA-Chromosome.jpg
- https://thumbs.dreamstime.com/z/human-cell-chromosome-telomere-repeating-sequence-double-stranded-dna-located-ends-chromosomes-each-time-36989577.jpg
- https://41.media.tumblr.com/e4c82ffee8f314dc8b45d77929d96a4c/tumblr_o2vm5rtT3k1v4ydioo1_r1_500.jpg
- http://b4fa.org/bioscience-in-brief/introduction-genes-crops/what-is-a-chromosome/
- http://www.bio.miami.edu/ecosummer/lectures/lec_genetics.html
- https://cnx.org/resources/647d3546c5b7f5cb57b87eda7bfb1b3599e6f231/0321_DNA_Macrostructure.jpg
Those images don't make much sense to me; with some images it's like the strands connect to the centromere of the chromosome, in others (like the dreamstime image), one section of the chromosome (2 arms, I suppose one long arm -p- and one short one -q- ) seem to be part of the strand alltogether, which makes much more sense to me and is more in line with what I read about the DNA replication/splitting (see https://en.wikipedia.org/wiki/DNA#/media/File:DNA_replication_en.svg ) It also seems to be in line of what I read on the miami page (that the chromosome are basically centromeres on which a clone of a section of the DNA strand (=chromosome) is made. So basically, the other long and short arm (a.k.a. chromatid) branching off from the centromere isn't strictly seen useful for the DNA, it can just be viewed as an temporary "add-on", and just a clone of a section of the main DNA strand.
I'm wondering whether someone can describe all this better on the chromosome page, and make an image that makes all this clear. The image could for instance be made based on https://en.wikipedia.org/wiki/DNA#/media/File:DNA_replication_en.svg but show the entire DNA strand including all chromosomes (so basically, just make a long DNA strand, and then mark the 23 chromosome lengths with brackets. The approximate lengths are mentioned at Chromosome#Human_chromosomes; basically:
- first 2 chromosomes= 8%
- chromosomes 3, 4, 5 = 6%
- chromosomes 6, 7, 8 = 5%
- chromosomes 9, 10, 11, 12 = 4%
- chromosomes 13, 14, 15, 16= 3%
- chromosomes 17, 18, 19, 20, 21= 2%
- chromosome 23= 5% or 2%
Also mark what's the p and q section of the chromosome, and indicate the centromere with a circle. I'm assuming you may want to use more colors for the nucleotid bars then in the DNA_replication_EN image, as I'm assuming all nucleotid bars need to be shown, see https://commons.wikimedia.org/wiki/File:Difference_DNA_RNA-EN.svg That is off course if it's possible to show the nucleotids at all (I'm assuming this might not be the case, as the image would then need to be impossibly long (as there are 6 000 000 000 nucleotids/base pairs). Even just showing a single gene wouldn't be possible if you were to have the image show any nucleotids (as each gene has about 260870 nucleotids -averaged out ie 6 billion/23000 genes-).
KVDP (talk) 09:18, 5 July 2016 (UTC)
There's been a new article regarding structural variation in the human genome, which is already covered in this article as a section while the main topic of Structural variation also has its own article. Just a pointer if anyone wishes to merge/expand or work with any of these... Ciridae (talk) 14:18, 28 August 2016 (UTC)
External links modified
Hello fellow Wikipedians,
I have just modified 2 external links on Human genome. Please take a moment to review my edit. If you have any questions, or need the bot to ignore the links, or the page altogether, please visit this simple FaQ for additional information. I made the following changes:
- Added archive https://web.archive.org/web/20100719235548/http://www.ncbi.nlm.nih.gov/About/primer/mapping.html to https://www.ncbi.nlm.nih.gov/About/primer/mapping.html
- Added archive https://web.archive.org/web/20150907140051/http://genome.wellcome.ac.uk/doc_WTD020876.html to http://genome.wellcome.ac.uk/doc_WTD020876.html
When you have finished reviewing my changes, you may follow the instructions on the template below to fix any issues with the URLs.
This message was posted before February 2018. After February 2018, "External links modified" talk page sections are no longer generated or monitored by InternetArchiveBot. No special action is required regarding these talk page notices, other than regular verification using the archive tool instructions below. Editors have permission to delete these "External links modified" talk page sections if they want to de-clutter talk pages, but see the RfC before doing mass systematic removals. This message is updated dynamically through the template {{source check}} (last update: 5 June 2024).
- If you have discovered URLs which were erroneously considered dead by the bot, you can report them with this tool.
- If you found an error with any archives or the URLs themselves, you can fix them with this tool.
Cheers.—InternetArchiveBot (Report bug) 14:00, 8 November 2017 (UTC)
Mutation rate
I tend to agree, somewhat, with the IP editor on this section. It is not up to snuff. It starts by citing creationist Michael Behe as source for mutation rate, but he is too self-serving in selecting data to either satisfy his agenda or serve as a strawman to be considered a WP:RS for a scientific article. Next it gives figure for genetic variation citing nothing more specific than "Researchers". There then follows a set of approximation calculations that appear to be the work of an editor, not derived from a source - basically everything from "Dividing that number . . ." to ". . . don't add up." looks like it is an unsourced, editor-generated calculation and conclusion. The next sentence, "Furthermore, Recent studies . . . new evolutionary time scale." doesn't really make sense - I know what it is trying to say because I read the cited Calloway piece, but it is a grammatical trainwreck and a word soup that is likely to be unintelligible to the naive reader. Then the Israeli specimens are thrown in as compounding the problem with regard to the human migration timeline, with no indication given why this is the case, and also without bringing it back to the subject being discussed (mutation rate, not migration timeline). I can certainly see why the IP found fault with this section. Agricolae (talk) 16:15, 25 June 2018 (UTC)
- Me too. There are multiple problems with the section. Among other things, it doesn't distinguish between the raw mutation rate (which can best be assessed by whole genome sequencing of parent-offspring trios, as has been done for a number of families recently), and the rate of accepted point mutations, such as would be assessed by comparing the genomes of two closely related species such as humans and chimpanzees. The Behe citation is also a concern, as Agricolae notes, and there are a number of instances of shooting from the hip, including using "apes" to refer specifically to chimpanzees, and miscalculating the number of bases in the human genome by a factor of 5-10. I'm not at all sure what point is being made in the end of the paragraph. I'm going to delete it again. If we want a discussion of mutation rate in this article, it should really be re-written from scratch. But a better place to contribute material on mutation rate would probably be in the article on mutation rate, which is already considerably better than the paragraph in this article. Jbening (talk) 23:08, 25 June 2018 (UTC)
Publication Date.
OK: this article states that the initial draft was published on the 12th February, 2001.
However … ?
The publication date is given as the 15th February: in the On This Day article for February 15th.
Is it possible we can get the wrong one, corrected?
Thanks
Cuddy2977 (talk) 14:33, 3 February 2019 (UTC)
- Yes--the date given in this article was that of the press release, which preceded actual publication. I've fixed it. Jbening (talk) 16:36, 3 February 2019 (UTC)
- My own view is that the precise day is unnecessary detail. Agricolae (talk) 21:01, 3 February 2019 (UTC)
Reversion
I undid this edit for a couple of reasons:
- Saying genomes were sequenced for $200 per person is misleading, because as the cited article explains, the company did that at a loss, so the true cost is still >$200.
- We can't say that estimates of the number of human genes went from 100,000 to 20,000 then up to >46,000, as the first two numbers are for protein-coding genes, whereas >46,000 includes RNA genes.
I apologise for the hasty wholesale reversion, which undid some good changes along with the disputed bits. Adrian J. Hunter(talk•contribs) 05:31, 31 May 2019 (UTC)
- Regarding these edits, I think the material on 'complete' genome sequencing needs a more nuanced approach. The human genome is usually spoken of as complete, even though there is some repetitive DNA and other hard-to-get-at regions that have not been fully spanned. You have to go pretty deep in the weeds to find a source that discusses the distinction between 'complete' and complete. I think more confusion than precision is likely if we insist on a perspective that while linguistically accurate is different than common usage not only in the popular scientific press, which routinely reports the 'complete' (really meaning 'sequenced with high fold coverage') genome sequencing of all kinds of species, and also in the scientific research community itself where it is spoken of more like 'complete (not really complete, but you know what we mean)' than incomplete. As written it seems almost to be an expression of a pet peeve about near-universal misuse of 'complete' when talking about genomes. I think it would be better to refer to it as complete while explaining that this doesn't really mean absolutely complete, than to state that it is not complete nor are any sequenced genomes in contrast to the way it is always reported and spoken of. Agricolae (talk) 13:18, 1 June 2019 (UTC)
- It's probably worth keeping a statement about completeness in the lede; most non-specialists I've talked with on the subject are surprised to hear that there are "degrees of completeness", even within the particular genome being sequenced. Their assumption is generally that we have a computer file which contains the entirety of binary DNA information, with no gaps. The general reaction to the probabilistic and overlap coverage description is "But that's not complete!" Tarl N. (discuss) 19:24, 1 June 2019 (UTC)
- I am not saying we shouldn't address completeness - it is a distinction worth making, but it needs to be nuanced. What I am not comfortable with is the recent edit. The original text was:
- "Completion of the Human Genome Project Sequence was published in 2004.[1] The human genome was the first of all vertebrates to be completely sequenced. As of 2012, thousands of human genomes have been completely sequenced, . . . "
- The editor was correct that this is problematic because it doesn't make clear what is meant by 'complete', that it doesn't mean the same as literally complete. However the new text:
- " . . . a more polished version published in 2004.[1] As of mid-2019, no human genome (nor any vertebrate genome) has been completely sequenced. "
- This goes the other direction. It was referred to then and afterwards as 'complete', even though everyone involved knew full well that this did not mean literally complete. This usage continues with the reporting to the 'complete' genome of the white tiger, even though it had more than a thousand gaps, and the 'complete' genome of the mandarin orange, even though it too had missing sequence. Simply put, 'complete' when talking about genomes, does not mean the same thing: 'complete genome sequence' is a term of art referring to sequencing the genome with high-fold coverage, even though some small regions of repetitive and 'unsequenceable' DNA remain unknown. We could even give a couple of sentences explicitly explaining the nature of the 'missing' sequence - I remember seeing a few years back an 'update' in Science or Nature with an overview of what types of DNA were still missing and interviewing a group using PacBio sequencing to try to close some of the gaps, so I know there is citable material out there to write such a summary. One way or another, I think we want to get across the point that it is referred to as complete (though it technically isn't), rather than just saying that no genome has ever been completely sequenced. Agricolae (talk) 20:16, 1 June 2019 (UTC)
- Oops, we already do describe this, just somewhere else in the article. I just think the recent edit went to far and obscures meaning rather than clarifying it. Agricolae (talk) 20:18, 1 June 2019 (UTC)
- OK, so I returned to the earlier text then modified to make the status explicit. As always, linguistic improvement is encouraged but I would rather we avoid the 'no genome has ever been completely sequenced' phrasing which, while literally true, doesn't match with typical usage. Agricolae (talk) 15:16, 2 June 2019 (UTC)
- I am not saying we shouldn't address completeness - it is a distinction worth making, but it needs to be nuanced. What I am not comfortable with is the recent edit. The original text was:
- It's probably worth keeping a statement about completeness in the lede; most non-specialists I've talked with on the subject are surprised to hear that there are "degrees of completeness", even within the particular genome being sequenced. Their assumption is generally that we have a computer file which contains the entirety of binary DNA information, with no gaps. The general reaction to the probabilistic and overlap coverage description is "But that's not complete!" Tarl N. (discuss) 19:24, 1 June 2019 (UTC)
References
Current list of human protein-coding genes
I know we have sub-articles on this with partial lists by chromosome, but there's now a complete list in the pages below in the event anyone is interested. I unfortunately couldn't add more information columns to those wikitables since I was running right up against the page size limit on both pages and wanted to split the list across as few pages as possible. Seppi333 (Insert 2¢) 19:28, 2 November 2019 (UTC)
- Wikipedia:WikiProject Molecular Biology/Molecular and Cell Biology/Human protein-coding genes1
- Wikipedia:WikiProject Molecular Biology/Molecular and Cell Biology/Human protein-coding genes2
july 2020 NIH X CHROMASOME RESULT
id like to add this to the article.
should i ?
and whare ?
- Not sure this is really the best place for it - the result is really more about the process of completing the sequence than it is about the genome itself. I would suggest: Human Genome Project#State of completion (and it would be better to cite the Nature paper than the press releases). Agricolae (talk) 21:07, 19 July 2020 (UTC)
Number of genes?
The article says
- As genome sequence quality and the methods for identifying protein-coding genes improved,[9] the count of recognized protein-coding genes dropped to 19,000-20,000.[12] However, a fuller understanding of the role played by sequences that do not encode proteins, but instead express regulatory RNA, has raised the total number of genes to at least 46,831,[13] plus another 2300 micro-RNA genes.[14]
but also says
- The haploid human genome (23 chromosomes) is about 3 billion base pairs long and contains around 30,000 genes.[29]
Which is it, or are both numbers, properly understood, correct? —WWoods (talk) 18:54, 14 November 2020 (UTC)
- It depends on what you count as a gene, and how long ago the analysis was done. That the 46k number comes from an article with the sub-headline: "The new estimate is based on a broader definition of just what a gene is". The second is more vague and generic, and it is unclear if it is rejecting the new definition of a gene offered by the other analysis, or if this is simply 'old data', that though the page has been updated as recently as this August, this particular datum is of older vintage. I think we need to see what sources from the past year and a half are saying about the gene count. Without that, the 46,831 number just represents one paper's conclusion that the definition of a gene should be changed, and what number that would produce, but has the field accepted this adjustment? Even if they have followed this thinking, the number is overly precise given that it results from making a whole lot of individual calls over whether each site is a gene or not. I think we should either present it less precisely as 'more than 46800' and likewise not present this redefinition of the gene as the recent 'new understanding' if it is just one paper's position - i.e. we should use conditional language, 'if the view of a gene is expanded, . . . ' or else we should use more descriptive language to refer to the precise number 'a recent analysis arguing the definition of a gene should be expanded concluded. . . .' so it is clear this is a single analysis with a specific set of assumptions. Agricolae (talk) 19:20, 14 November 2020 (UTC)
Junk DNA
90% of the human genome is junk but "junk" is only mentioned once in the article. That needs to be fixed. Genome42 (talk) 20:40, 27 July 2022 (UTC)
- Yeah, it needs to be removed. 'Junk' DNA is a concept defined by what it isn't, and is no longer considered a useful distinction given the diverse types of functional (in some cases critically important) and non-functional DNA that is included in this catch-all term. I have removed it. Agricolae (talk) 22:40, 27 July 2022 (UTC)
- There is solid evidence that 90% of or genome is junk. There will soon be a separate Wikipedia article devoted to this topic. You need to read up on this topic. You might want to start with one of my blog posts.
- "Five Things You Should Know if You Want to Participate in the Junk DNA Debate"
- https://sandwalk.blogspot.com/2013/07/five-things-you-should-know-if-you-want.html Genome42 (talk) 20:16, 29 July 2022 (UTC)
- This is not the 1980s, when if DNA didn't encode proteins then we just threw up our hands and dismissively called it 'junk'. A promoter is not 'junk'. A centromere is not 'junk'. rDNA is not 'junk'. And most importantly, these don't belong in the same heterogeneous category with each other, let alone in the same artificial category with LINEs, pseudogenes, viral insertions, etc. We already mention so-called 'junk' DNA in the article, just not using the inaccurate term for it - "Human genomes include both protein-coding DNA genes and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences." There is little benefit in ignoring this diversity, lumping it all together under a heterogeneous catch-all term of 'all of the various different things that aren't X'. More useful would be to break out the unhelpful '90%' figure into its distinct constituent parts, which data should be in the 2022 Science Special Issue (currently behind a paywall for me). Agricolae (talk) 20:57, 29 July 2022 (UTC)
- Just a followup, elsewhere you have defined junk DNA as "DNA that can be deleted from the genome without affecting the fitness of the individual or the species". I would suggest that no piece of the human DNA has been characterized to the level at which we would know this for certain, and it would be unethical to carry out the test. It amounts to a 'we don't know of a benefit so there must not be one' argument. Given that pseudogenes can provide a venue for variation that can then be incorporated back into the transcriptome through gene conversion or reactiveation, that we all have at least one retrotransposed mRNAs in our genome that is a fully-functionaltranscribed protein because it randomly inserted next to a promoter, or a promoter randomly inserted next to it, and that LINEs allow for unequal crossing over and consequent gene duplication, specialization and diversification, I don't know how you point to a particular piece of DNA and say that having it can't possibly ever provide a fitness advantage. In your blog you point to the range of difference in genome size, but there are genomes stripped down to the point where they have next to no 'junk DNA' - if they can, so could we, but we don't - it has been argued that this is evidence for a functional benefit to all that misnamed 'junk', even if we don't know how such a fitness benefit might come about. And all of that doesn't get around the fundamental problem that 'junk' DNA is a heterogeneous category, defined by what it isn't, and like Tolstoy's unhappy families, all unhappy in their own different way, nothing more than a catchall for whatever things that are not something else, a combination of a linguistic holdover from a previous time and an expression of current imperfect knowledge and imagination. Even if it really is useless, and we certainly can't be sure that is the case, we are much better off talking individually about pseudogenes, LINEs, etc., rather than about an artificial category including all of these disparate things, each unhappy for its own particular reason. We are usually better off describing green, and blue, and yellow, rather than creating a page for 'colours that aren't red'.Agricolae (talk) 00:35, 30 July 2022 (UTC)
- Much of what you have just written is scientifically incorrect, misleading, or logically flawed (sometimes all three). But your lack of knowledge of the scientific controversy over junk DNA isn't really the point. The point is that there are many highly respected and intelligent scientists who say that most of our genome consists of junk DNA. You may not agree with them but you are wrong to use your power as a Wikipedia editor to impose your personal opinion on a Wikipedia article. It's ridiculous to censure any mention of "junk DNA" when the term is used in lots of other Wikipedia articles and is part of a very interesting and ongoing controversy about the content of the human genome.
- I've posted your comments on my blog where we can discuss all the bits that need correcting.
- Wikipedia blocks any mention of junk DNA in the "Human genome" article
- https://sandwalk.blogspot.com/2022/07/wikipedia-blocks-any-mention-of-junk.html Genome42 (talk) 15:40, 30 July 2022 (UTC)
- Do what you want on your personal playground - that has nothing to do with Wikipedia. Here I would rather discuss why it is important to have a classification for what remains a Tolstoyan 'unhappy families' category that combines didfferent types of DNA with different origins, regulation and hypothesized evolutionary contribution(s). It is a loss of information, bordering on intentional obfuscation, to take defined genomic proportions of LINEs, pseudogenes, SINEs, etc., etc., and simply report that as a combined "90% of the genome is 'other stuff'" instead of being specific. Agricolae (talk) 02:27, 31 July 2022 (UTC)
- Me writing on wikipedia is no different than you writing on wikipedia, so you are no less treating this as your own personal playground than any of us here. Several others of whom are actual experts in their respective fields. You have not provided any real evidence against both the reality of the existence of junk DNA(DNA with no function), nor have you provided any substantive rebuttal to the many articles that have already been by offered by Genome42 in an earlier post.
- The fact is that there is only good evidence for functionality for a small percentage of the human genome(essentially all papers that argue to the contrary are speculating by extrapolating from a small handful of examples that all other genomic elements of the type studied-be they retrotransposons, alternatively spliced genes, pseudogenes or what have you-could hypthetically, in principle, function in a similar way.) And there are many good evidence-based reasons for thinking the remaining is nonfunctional junk DNA that got left there as a product of evolution. This hypothesis has the advantage that it both incorporates what we know from direct experiments of how transcription factors interact with DNA, and it accounts for the large variations in genome size between different species. And unlike the "it's all(or almost all) functional" hypothesis, it's also consistent with what we know from population genetics about the relationships between genome size, mutation rate, population size, and the resulting mutational load, not to mention the degree of conservation on both sequence and length from interspecies genome comparisons.
- And then it's a better scientific hypothesis because it takes very little to actually reject nonfunctional junk as a nullhypothesis, contrary to the hypothesis that it's all functional in a way we have yet to show because the latter functionality hypothesis can admit to an endless number of rationalizations that the experiment just didn't include some obscure effect that would have brought out the functional effect consequences.
- It is also a demonstrable historical falsehood to say that all non-coding DNA was once thought to be nonfunctional junk. Reasons have been given already for why your policing the article on the human genome against mentioning the concept of junk DNA is both in conflict with Wikipedia policy, and scientifically and historically unjustified.
- Even if you personally think that junkDNA is wrong, this isn't some sort of pseudoscience like homeopathy, clairvoyance, or remote viewing. The subject of junk-vs-function and it's relationship to coding-vs-noncoding is a a field being actively studied in established scientific institutions and university departments, and by qualified experts from a range of fields ranging from molecular biology, through evolution, to population genetics. It is a debate about the relative proportions in the genomes of different organisms, not about whether there even is such a thing as junk DNA. It is disconcerting that you are here personally stifling exposure to a legitimate scientific subject where there is still an ongoing, and increasing amount of scientific debate, and that Wikipedia through your actions are here becoming a vessel for misleading the public and upcoming scientists, rather than to serve as informative to the latest developments in the field. Rumraket38 (talk) 13:44, 31 July 2022 (UTC)
- And I just have to add that you are just flatly mistaken and seem to be laboring under a severe misapprehension if you think we are saying all non-coding DNA is nonfunctional junk DNA.
- If you think those of us who are advocating a mention and discussion of the junk DNA concept in this article on the human genome are saying anything that doesn't code for protein is automatically junk then you have completely misunderstood us. Rumraket38 (talk) 13:51, 31 July 2022 (UTC)
- Do what you want on your personal playground - that has nothing to do with Wikipedia. Here I would rather discuss why it is important to have a classification for what remains a Tolstoyan 'unhappy families' category that combines didfferent types of DNA with different origins, regulation and hypothesized evolutionary contribution(s). It is a loss of information, bordering on intentional obfuscation, to take defined genomic proportions of LINEs, pseudogenes, SINEs, etc., etc., and simply report that as a combined "90% of the genome is 'other stuff'" instead of being specific. Agricolae (talk) 02:27, 31 July 2022 (UTC)
- At some point Agricolae needs to recognize that the opinion Genome42 expressed is not just a minority view held by a few cranks. I wouldn't go so far as to say that it's the majority opinion of protein chemists, but it may be. Is there a single protein chemist that anyone can cite (full reference please) who disagree with Genome42? Note also that Genome42's qualifications are very well known and easily checked: his textbook is one of the most influencial biochemistry textbooks in use today. Unfortunately I have not had any success trying to discover what qualifications his opponents have. Some non-biochemists may agree that junk DNA is passé, maybe even a majority, but there are certainly some geneticists and molecular biologists who do not. Dan Graur, for example, has expressed himself very forcefully on the the subject, and so did Sydney Brenner when he was alive. This argument will not go away. Athel cb (talk) 18:08, 30 July 2022 (UTC)
- @Athel cb I'm unaware of any but who knows, there are odd individuals out there. I'd rather ask, can you find an enzymologist who would agree with ENCODE's apparent assertion that only functional bits of DNA get transcribed? That would indeed suggest some magic powers for RNA transcriptase. 146.115.157.23 (talk) 15:06, 2 August 2022 (UTC)
- I'm not aware of any, but they may exist. I'd be surprised if anyone who understands the subject would would agree with ENCODE's apparent assertion that only functional bits of DNA get transcribed. It strikes me as utterly absurd (and close to magic). However, if such people exist it's Agricolae's job to find them and cite them. Repeatedly posting a dogmatic opinion as if it's established fact needs to stop. Athel cb (talk) 15:33, 2 August 2022 (UTC)
- @Athel cb I'm unaware of any but who knows, there are odd individuals out there. I'd rather ask, can you find an enzymologist who would agree with ENCODE's apparent assertion that only functional bits of DNA get transcribed? That would indeed suggest some magic powers for RNA transcriptase. 146.115.157.23 (talk) 15:06, 2 August 2022 (UTC)
- I have to side with both Genome42 and Athel cb here. I have been following the junk-DNA controversy for over a decade now and it is in NO way a fringe theory. It is a legitimate scientific controversy that is being actively published on and researched. The claim that it is fringe that there is junk DNA, or that it is settled that there is no junk DNA is empirically and historically an outright falsehood, and there is substantial support for both the reality that junk DNA exists, and that a significant fraction of the human genome is in fact junk DNA.
- I think Agricolae needs to familiarize itself with the considerable literature that has already been cited earlier by Genome42 as pretty much all of the arguments brought up by Agricolae are addressed at length in those papers. Rumraket38 (talk) 22:35, 30 July 2022 (UTC)
- Let me add my agreement to the comments by Laurence Moran (Genome42), Athel Cornish-Bowden (Athel cb), and Rumraket. There is a dramatic contrast between the views of almost all researchers on molecular evolution, and the views of other genomicists and molecular biologist. The former understand the evidence that most of our genome is "junk DNA". The latter are basing themselves mostly on recent hearsay. This is a sad situation and a major disagreement, and it is outrageous that Wikipedia does not allow even a mention of it in this article. Felsenst (talk) 23:41, 30 July 2022 (UTC) (Joe Felsenstein, Emeritus Professor, Department of Genome Sciences, University of Washington, Seattle (also Department of Biology).
- @Agricolae As someone who was a practicing molecular biologist in the 80s and beyond, your response puzzles me. I never witnessed anyone call all non-coding DNA junk, or ignored functional categories like rRNA, tRNA, centromeres, promotors, origins of replication, and additional sequence specific sites that bind regulatory proteins. If they had, they would have been checked by more knowledgeable people. So your comment begins with an ahistorical foundation. Rather, we spoke of junk regarding the mysterious fact that there was so much more DNA than had any apparent function, than appeared to have any genetic significance. With the ENCODE boondoggle (and a few misadventures beforehand), we saw an abuse of the meaning of function to mean "something we could measure in a biochemical assay", or close to it. Data scientists who seem to have skipped biochemistry and genetics didn't understand that we expect processes like transcription (and even translation) to be "leaky". The biochemical machines that make RNA copies aren't smart enough to know that something that a random occurrence of a promoter isn't neighboring something useful. That assertion bothers some people, but they are almost never biochemists and never enzymologists. I'm guessing you're neither. Before attempting to speak with authority, please consult some enzymologists and biochemists.
- One could go on with the fact that transposable elements that become non-function are expected to proliferate and leave behind non-functional DNA. Population Genetics reveals that there is, to the best approximations, inadequate selection to clear them. Overall, the science is clear, and you're not helping. 146.115.157.23 (talk) 14:44, 2 August 2022 (UTC)
- Just a followup, elsewhere you have defined junk DNA as "DNA that can be deleted from the genome without affecting the fitness of the individual or the species". I would suggest that no piece of the human DNA has been characterized to the level at which we would know this for certain, and it would be unethical to carry out the test. It amounts to a 'we don't know of a benefit so there must not be one' argument. Given that pseudogenes can provide a venue for variation that can then be incorporated back into the transcriptome through gene conversion or reactiveation, that we all have at least one retrotransposed mRNAs in our genome that is a fully-functionaltranscribed protein because it randomly inserted next to a promoter, or a promoter randomly inserted next to it, and that LINEs allow for unequal crossing over and consequent gene duplication, specialization and diversification, I don't know how you point to a particular piece of DNA and say that having it can't possibly ever provide a fitness advantage. In your blog you point to the range of difference in genome size, but there are genomes stripped down to the point where they have next to no 'junk DNA' - if they can, so could we, but we don't - it has been argued that this is evidence for a functional benefit to all that misnamed 'junk', even if we don't know how such a fitness benefit might come about. And all of that doesn't get around the fundamental problem that 'junk' DNA is a heterogeneous category, defined by what it isn't, and like Tolstoy's unhappy families, all unhappy in their own different way, nothing more than a catchall for whatever things that are not something else, a combination of a linguistic holdover from a previous time and an expression of current imperfect knowledge and imagination. Even if it really is useless, and we certainly can't be sure that is the case, we are much better off talking individually about pseudogenes, LINEs, etc., rather than about an artificial category including all of these disparate things, each unhappy for its own particular reason. We are usually better off describing green, and blue, and yellow, rather than creating a page for 'colours that aren't red'.Agricolae (talk) 00:35, 30 July 2022 (UTC)
- This is not the 1980s, when if DNA didn't encode proteins then we just threw up our hands and dismissively called it 'junk'. A promoter is not 'junk'. A centromere is not 'junk'. rDNA is not 'junk'. And most importantly, these don't belong in the same heterogeneous category with each other, let alone in the same artificial category with LINEs, pseudogenes, viral insertions, etc. We already mention so-called 'junk' DNA in the article, just not using the inaccurate term for it - "Human genomes include both protein-coding DNA genes and various types of DNA that does not encode proteins. The latter is a diverse category that includes DNA coding for non-translated RNA, such as that for ribosomal RNA, transfer RNA, ribozymes, small nuclear RNAs, and several types of regulatory RNAs. It also includes promoters and their associated gene-regulatory elements, DNA playing structural and replicatory roles, such as scaffolding regions, telomeres, centromeres, and origins of replication, plus large numbers of transposable elements, inserted viral DNA, non-functional pseudogenes and simple, highly-repetitive sequences." There is little benefit in ignoring this diversity, lumping it all together under a heterogeneous catch-all term of 'all of the various different things that aren't X'. More useful would be to break out the unhelpful '90%' figure into its distinct constituent parts, which data should be in the 2022 Science Special Issue (currently behind a paywall for me). Agricolae (talk) 20:57, 29 July 2022 (UTC)
I think this is a sematic issue. It is clear that at least some DNA that was formerly called junk has a function. Neverthelss, there remains a large percentage of DNA that has no apparent function, at least no function that has yet been identified. Finally some DNA may not have an immediate function in living organisms, but may still play an evolutionary role.[1] The term junk DNA is widely used in the literature, hence I think it should at least be mentioned with all the above caveats. Boghog (talk) 19:57, 30 July 2022 (UTC)
- We have indeed found a few select categories of functional DNA that were not know in the 80s. Those don't include tRNA, rRNA, promoters, operators, origins of replication, or centromeres or telomers. The amount of the genome with newly discovered functional elements is small. The fact remains that well over 80% is junk. What is clear is that there is surprising confusion over the meaning of junk, mostly among non-biologists or sloppy biologists who are guilty of gross over-simplifications. Sadly, such gross over-simplifications are sometimes promoted under the pretext of making science more accessible, but ultimately they do significant damage. 146.115.157.23 (talk) 14:55, 2 August 2022 (UTC)
References
- ^ Andolfatto P (October 2005). "Adaptive evolution of non-coding DNA in Drosophila". Nature. 437 (7062): 1149–52. doi:10.1038/nature04107. PMID 16237443. Lay summary in: "UCSD Study Shows 'Junk' DNA Has Evolutionary Importance". ScienceDaily. Rockville, MD. 20 October 2005.
- Yes, and take it further - these roles, these potential functions are different for each different type of so-called 'junk'. The hypothesized evolutionary role(s) of LINEs is/are different than the potential evolutionary role(s) of pseudogenes. We don't have an article for 'all of the different functional types of DNA with known roles' that tries to combine a discussion of promoters and coding sequence and rDNA and origins of replication, we discuss the individual roles of each specific type, yet 'junk DNA' is analogously heterogeneous and less helpful than similarly individualized discussion of the various components dismissively clustered in that 'everything else' pseudo-category. Agricolae (talk) 02:27, 31 July 2022 (UTC)
- OK, so you have a reference to someone who thought that some non-coding DNA had a function, but there is no argument about that. No evidence, however, from anyone with expertise in biochemistry or protein chemistry who thinks junk DNA is a discredited concept. Let's have a serious reference, please. Let's also take seriously Dan Graur's calculation (pp. 514-517 in his book Molecular and Genome Evolution[1]) that if all DNA is essential then a human parent needs to have an absolutely vast number of children to maintain a viable population. If Graur is wrong, then let's have an analysis that shows exactly where he went wrong (good luck with that). While you're at it, let's have an explanation of why onions and lungfish need so much more DNA than humans. Of course, if you take the religious view that humans are the pinnacle of God's creation then you have no trouble with that, but few modern biologists will agree. Athel cb (talk) 07:10, 31 July 2022 (UTC)
- According to WP:NPOV, articles should include all significant points of view that are supported by reliable sources. While the concept of junk DNA is controversial,[2] it has at the very least significant minority and probably majority support within the sceintic community. There are numerous sources that support that a signficant fraction of our genome is junk.[3][4][5] Hence I strongly agree with Genome42, Athel cb, Rumraket38, and Felsenst that "junk DNA" should be mentioned in this article. Not doing so is a disservice to our readers. Boghog (talk) 08:09, 31 July 2022 (UTC)
References
- ^ Graur D (2016). Molecular and Genome Evolution. Sunderland, Massachusetts: Sinauer Sunderland. pp. 514–517. ISBN 978-1-60535-469-9.
- ^ Pena SD (2021). "An Overview of the Human Genome: Coding DNA and Non-Coding DNA". In Haddad LA (ed.). Human Genome Structure, Function and Clinical Considerations. Cham: Springer Nature. pp. 5–7. ISBN 978-3-03-073151-9.
- ^ Palazzo AF, Gregory TR (May 2014). "The case for junk DNA". PLoS Genetics. 10 (5): e1004351. doi:10.1371/journal.pgen.1004351. PMC 4014423. PMID 24809441.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Graur D (2017). "Rubbish DNA: the functionless fraction of the human genome." (PDF). Evolution of the Human Genome I. Tokyo: Springer. pp. 19–60. doi:10.1007/978-4-431-56603-8_2. ISBN 978-4-431-56603-8.
- ^ Graur D (July 2017). "An Upper Limit on the Functional Fraction of the Human Genome". Genome Biology and Evolution. 9 (7): 1880–1885. doi:10.1093/gbe/evx121. PMC 5570035. PMID 28854598.Lay summary in: Le Page M (17 July 2017). "At least 75 per cent of our DNA really is useless junk after all". NewScientist.
- I agree with Boghog; this is a semantic issue. The way to handle semantic issues is not to avoid the word in question. Instead, the solution is to define the word, or at least discuss its definition. IIUC a consensus is that "non-protein-coding DNA" is misleading, and there are other DNA categories and functions besides coding proteins. This suggests to me to use that consensus: "Junk DNA: noun: DNA which has no known function." Put disagreements on what qualifies as functional in the larger discussion. Eddyalumni (talk) 08:39, 31 July 2022 (UTC)
- Yes I think it's important to get the distinction right, thank you for that. Those of us who are advocating that this article on the human genome includes discussion of the concept of junk DNA and what proportion of it would merit the use of this term are definitely not saying that non-coding DNA is by definition nonfunctional. I would totally agree that is a mistaken view. But I also think that if one is careful to define one's terms, that the concept of DNA that doesn't have a function is a legitimate concept, and that a proper term for this is junk DNA, then one can in fact reference junk DNA in this article and provide a discussion of what proportion of the human genome corresponds to junk DNA(DNA that doesn't have a function regardless of whether it is coding or not). Rumraket38 (talk) 13:48, 31 July 2022 (UTC)
- This is not a "semantic" issue. It's a scientific issue. We want to know how much of the human genome is functional but there's no clear definition of "function" in the article. This is important because there are tables showing more than 15,000 lncRNA genes but no discussion of how these "genes" are defined. Are all transcipts functional? Of course they aren't; fewer than 500 lncRNAs have actually been shown to have any evidence of function.
- Real genes have to be functional, otherwise they are pseudogenes, hypothetical genes, or mirages.
- In the section on "Regulatory DNA sequences" there's a statement that they make up 8% of the genome but no attempt to define which of the presumptive regulatory sequences are functional. (Most aren't.) The article then goes on to say that according to ENCODE the regulatory sequences might occupy 20% t0 40% of the genome but that's ridiculous. No knowledgeable scientist has come up with a reasonable definition of function that would include that amount of regulatory sequence.
- In the section on transposons, the article currently implies that half of our genome is composed of non-functional relics of transposons without mentioning how we know that they are non-functional. (They are.) For some strange reason, the former editors want to avoid calling this DNA "junk" but that doesn't make sense since the definition of junk DNA is non-functional DNA.
- The point is that there's a considerable amount of hypocrisy on display here. Some people are demanding a rigorous definition of junk (i.e. not function) but they aren't as rigorous when it comes to describing function. This is where "semantics" is overruling science and it needs to stop. Genome42 (talk) 15:45, 31 July 2022 (UTC)
- I am not demanding anything. And I agree with you 100% that we need to be rigorous when defining function and that other sections of this article need editing (e.g., "Regulatory DNA sequences", "transposons"). Finally, I think we now have a strong consensus on this talk page that "junk DNA" should be mentioned in this article. Boghog (talk) 16:27, 31 July 2022 (UTC)
Noncoding DNA (ncDNA)
I suggest consideration of the following reference wherein one finds "Despite popular arguments for functionality of most, if not all, of these transcripts, genome-wide analysis of selective constraints indicates that most of the produced RNA are junk." Sorry I'm not registering or formatting the reference correctly. The point is that the text seems to assert function to most transcribed DNA but this is not a well accepted contemporary opinion. Note this is the same author cited in ref 58 in this section.
Alexander F.Palazzo Eugene V.Koonin (2020) Cell Volume 183, Issue 5], 25 November 2020, Pages 1151-1161 https://doi.org/10.1016/j.cell.2020.09.047 146.115.157.23 (talk) 15:39, 2 August 2022 (UTC)
- The heart of the controversy is that some have argued that if DNA is transcribed, it must be functional. This review article however presents evidence that the much of the produced RNA is junk,[1] and therefore by definition, the corresonding DNA must also be junk. It is a good source, particularily since it is secondary (a review article) and also not hiding behing a paywall. Boghog (talk) 21:17, 2 August 2022 (UTC)
The article contains two tables that list tens of thousands of noncoding genes. Many of us are highly skeptical of that estimate for the reasons that my friend and colleague describes in his paper with Eugene Koonin and in another paper with his graduate student [2]. The tables in the Wikipedia article are very misleading. The big one should probably be deleted since it contains obviously false information such as the number of ribosomal RNA genes. The small one needs caveats in the main body of the article pointing out that the gene estimates are not supported by direct evidence of function.
In addition to the Palazzo and Koonin (2020) article, there are many other earlier articles that make the same point. Several were published right after the misleading ENCODE publicity campaign in 2012.Genome42 (talk) 21:55, 2 August 2022 (UTC)
- This work suggests a lot of IncRNA still goes to ribosome, even though protein is useless. https://twitter.com/RNAcentral/status/1588204509918003202?s=19 2A00:1370:8184:1765:5A9F:E8F2:BF58:135B (talk) 01:20, 7 November 2022 (UTC)
- ^ Palazzo AF, Koonin EV (November 2020). "Functional Long Non-coding RNAs Evolve from Junk Transcripts". Cell. 183 (5): 1151–1161. doi:10.1016/j.cell.2020.09.047. PMID 33068526. S2CID 222815635.
- ^ Palazzo AF, and Lee ES (2015). "Non-coding RNA: what is functional and what is junk?". Frontiers in genetics. 6: 1–11. doi:10.3389/fgene.2015.00002.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
Featured picture scheduled for POTD
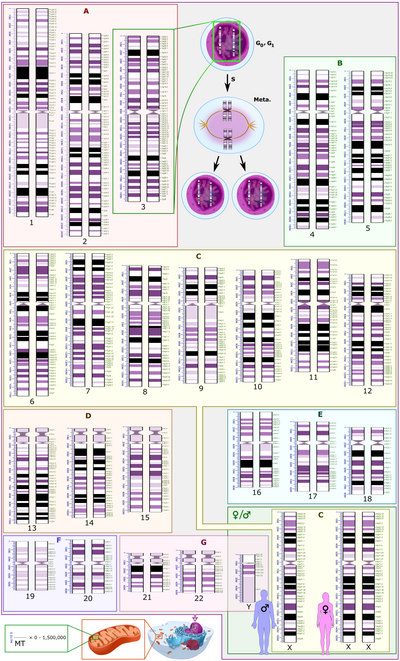
Hello! This is to let editors know that File:Human karyotype_with_bands_and_sub-bands.png, a featured picture used in this article, has been selected as the English Wikipedia's picture of the day (POTD) for February 20, 2023. A preview of the POTD is displayed below and can be edited at Template:POTD/2023-02-20. For the greater benefit of readers, any potential improvements or maintenance that could benefit the quality of this article should be done before its scheduled appearance on the Main Page. If you have any concerns, please place a message at Wikipedia talk:Picture of the day. Thank you! — Amakuru (talk) 22:55, 8 February 2023 (UTC)
- That picture is outdated, https://en.wikipedia.org/wiki/File_talk:Human_karyotype_with_bands_and_sub-bands.png 2A00:1370:8184:1CE9:B087:1D85:2341:205A (talk) 07:36, 7 March 2023 (UTC)
|
The human genome is the complete set of nucleic acid sequences for humans, encoded as DNA within the 23 chromosome pairs in cell nuclei and in small DNA molecules found within mitochondria. This includes both protein-coding DNA sequences and various types of DNA that does not encode proteins. This schematic representation of the human diploid karyotype shows the organization of the human genome into chromosomes, as well as annotated bands and sub-bands as seen on G banding. The diagram shows both the female (XX) and male (XY) versions of the 23rd chromosome pair. Chromosomal changes during the cell cycle are displayed at the top center. The human mitochondrial genome is shown to scale at the bottom left. Diagram credit: Mikael Häggström
Recently featured:
|
