Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency.[1] Nucleases are also extensively used in molecular cloning.[2]
There are two primary classifications based on the locus of activity. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the middle of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA.[2]
History
[edit]In the late 1960s, scientists Stuart Linn and Werner Arber isolated examples of the two types of enzymes responsible for phage growth restriction in Escherichia coli (E. coli) bacteria.[3][4] One of these enzymes added a methyl group to the DNA, generating methylated DNA, while the other cleaved unmethylated DNA at a wide variety of locations along the length of the molecule. The first type of enzyme was called a "methylase" and the other a "restriction nuclease". These enzymatic tools were important to scientists who were gathering the tools needed to "cut and paste" DNA molecules. What was then needed was a tool that would cut DNA at specific sites, rather than at random sites along the length of the molecule, so that scientists could cut DNA molecules in a predictable and reproducible way.
An important development came when H.O. Smith, K.W. Wilcox, and T.J. Kelly, working at Johns Hopkins University in 1968, isolated and characterized the first restriction nuclease whose functioning depended on a specific DNA nucleotide sequence. Working with Haemophilus influenzae bacteria, this group isolated an enzyme, called HindII, that always cut DNA molecules at a particular point within a specific sequence of six base pairs. They found that the HindII enzyme always cuts directly in the center of this sequence (between the 3rd and 4th base pairs).
Numerical Classification System
[edit]Most nucleases are classified by the Enzyme Commission number of the "Nomenclature Committee of the International Union of Biochemistry and Molecular Biology" as hydrolases (EC-number 3). The nucleases belong just like phosphodiesterase, lipase and phosphatase to the esterases (EC-number 3.1), a subgroup of the hydrolases. The esterases to which nucleases belong are classified with the EC-numbers 3.1.11 - EC-number 3.1.31.
Structure
[edit]Nuclease primary structure is by and large poorly conserved and minimally conserved at active sites, the surfaces of which primarily comprise acidic and basic amino acid residues. Nucleases can be classified into folding families.[5]

Site recognition
[edit]A nuclease must associate with a nucleic acid before it can cleave the molecule. That entails a degree of recognition. Nucleases variously employ both nonspecific and specific associations in their modes of recognition and binding. Both modes play important roles in living organisms, especially in DNA repair.[7]
Nonspecific endonucleases involved in DNA repair can scan DNA for target sequences or damage. Such a nuclease diffuses along DNA until it encounters a target, upon which the residues of its active site interact with the chemical groups of the DNA. In the case of endonucleases such as EcoRV, BamHI, and PvuII, this nonspecific binding involves electrostatic interactions between minimal surface area of the protein and the DNA. This weak association leaves the overall shape of the DNA undeformed, remaining in B-form.[7]
A site-specific nuclease forms far stronger associations by contrast. It draws DNA into the deep groove of its DNA-binding domain. This results in significant deformation of the DNA tertiary structure and is accomplished with a surfaces rich in basic (positively charged) residues. It engages in extensive electrostatic interaction with the DNA.[7]
Some nucleases involved in DNA repair exhibit partial sequence-specificity. However most are nonspecific, instead recognizing structural abnormalities produced in the DNA backbone by base pair mismatches.[7]
Structure specific nuclease
[edit]For details see flap endonuclease.
Sequence specific nuclease
[edit]| Enzyme | Source | Recognition Sequence | Cut |
|---|---|---|---|
| HindII | Haemophilus influenzae |
5'–GTYRAC–3' |
5'–GTY RAC–3' |
| R = A or G; Y = C or T | |||
There are more than 900 restriction enzymes, some sequence specific and some not, have been isolated from over 230 strains of bacteria since the initial discovery of HindII. These restriction enzymes generally have names that reflect their origin—The first letter of the name comes from the genus and the second two letters come from the species of the prokaryotic cell from which they were isolated. For example, EcoRI comes from Escherichia coli RY13 bacteria, while HindII comes from Haemophilus influenzae strain Rd. Numbers following the nuclease names indicate the order in which the enzymes were isolated from single strains of bacteria: EcoRI, EcoRII.
Endonucleases
[edit]A restriction endonuclease functions by "scanning" the length of a DNA molecule. Once it encounters its particular specific recognition sequence, it will bind to the DNA molecule and makes one cut in each of the two sugar-phosphate backbones. The positions of these two cuts, both in relation to each other, and to the recognition sequence itself, are determined by the identity of the restriction endonuclease. Different endonucleases yield different sets of cuts, but one endonuclease will always cut a particular base sequence the same way, no matter what DNA molecule it is acting on. Once the cuts have been made, the DNA molecule will break into fragments.
Staggered cutting
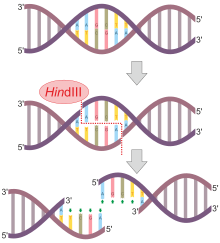
[edit]Not all restriction endonucleases cut symmetrically and leave blunt ends like HindII described above. Many endonucleases cleave the DNA backbones in positions that are not directly opposite each other, creating overhangs. For example, the nuclease EcoRI has the recognition sequence 5'—GAATTC—3'.
| Enzyme | Source | Recognition Sequence | Cut |
|---|---|---|---|
| HindIII | Haemophilus influenzae |
5'–AAGCTT–3'
3'–TTCGAA–5' |
5'–A AGCTT–3'
3'–TTCGA A–5' |
| EcoRI | Escherichia coli | 5'–GAATTC-3'
3'–CTTAAG–5' |
5'–G AATTC–3'
3'–CTTAA G–5' |
| BamHI | Bacillus amyloliquefaciens | 5'–GGATCC–3'
3'–CCTAGG–5' |
5'–G GATCC–3'
3'–CCTAG G–5' |
When the enzyme encounters this sequence, it cleaves each backbone between the G and the closest A base residues. Once the cuts have been made, the resulting fragments are held together only by the relatively weak hydrogen bonds that hold the complementary bases to each other. The weakness of these bonds allows the DNA fragments to separate from each other. Each resulting fragment has a protruding 5' end composed of unpaired bases. Other enzymes create cuts in the DNA backbone which result in protruding 3' ends. Protruding ends—both 3' and 5'—are sometimes called "sticky ends" because they tend to bond with complementary sequences of bases. In other words, if an unpaired length of bases 5'—AATT—3' encounters another unpaired length with the sequence 3'—TTAA—5' they will bond to each other—they are "sticky" for each other. Ligase enzyme is then used to join the phosphate backbones of the two molecules. The cellular origin, or even the species origin, of the sticky ends does not affect their stickiness. Any pair of complementary sequences will tend to bond, even if one of the sequences comes from a length of human DNA, and the other comes from a length of bacterial DNA. In fact, it is this quality of stickiness that allows production of recombinant DNA molecules, molecules which are composed of DNA from different sources, and which has given birth to the genetic engineering technology.
Role in nature
[edit]DNA repair
[edit]With all cells depending on DNA as the medium of genetic information, genetic quality control is an essential function of all organisms. DNA replication is an error prone process, and DNA molecules themselves are vulnerable to modification by many metabolic and environmental stressors. Ubiquitous examples include reactive oxygen species, near ultraviolet, and ionizing radiation. Many nucleases participate in DNA repair by recognizing damage sites and cleaving them from the surrounding DNA. These enzymes function independently or in complexes. Most nucleases involved in DNA repair are not sequence-specific. They recognize damage sites through deformation of double stranded DNA (dsDNA) secondary structure.[5]
Replication proofreading
[edit]During DNA replication, DNA polymerases elongate new strands of DNA against complementary template strands. Most DNA polymerases comprise two different enzymatic domains: a polymerase and a proofreading exonuclease. The polymerase elongates the new strand in the 5' → 3' direction. The exonuclease removes erroneous nucleotides from the same strand in the 3’ → 5’ direction. This exonuclease activity is essential for a DNA polymerase's ability to proofread. Deletions inactivating or removing these nucleases increase rates of mutation and mortality in affected microbes and cancer in mice.[8]
Halted replication fork
[edit]Many forms of DNA damage stop progression of the replication fork, causing the DNA polymerases and associated machinery to abandon the fork. It must then be processed by fork-specific proteins. The most notable is MUS81. Deletions of which causes UV or methylation damage sensitivity in yeast, in addition to meiotic defects.[5]
Okazaki fragment processing
[edit]A ubiquitous task in cells is the removal of Okazaki fragment RNA primers from replication. Most such primers are excised from newly synthesized lagging strand DNA by endonucleases of the family RNase H. In eukaryotes and in archaea, the flap endonuclease FEN1 also participates in the processing of Okazaki fragments.[5]
Mismatch repair
[edit]DNA mismatch repair in any given organism is effected by a suite of mismatch-specific endonucleases. In prokaryotes, this role is primarily filled by MutSLH and very short patch repair (VSP repair) associated proteins.
The MutSLH system (comprising MutS, MutL, and MutH) corrects point mutations and small turns. MutS recognizes and binds to mismatches, where it recruits MutL and MutH. MutL mediates the interaction between MutS and MutH, and enhances the endonucleasic activity of the latter. MutH recognizes hemimethylated 5'—GATC—3' sites and cleaves next to the G of the non-methylated strand (the more recently synthesized strand).
VSP repair is initiated by the endonuclease Vsr. It corrects a specific T/G mismatch caused by the spontaneous deamination of methylated cytosines to thymines. Vsr recognizes the sequence 5'—CTWGG—3',
where it nicks the DNA strand on the 5' side of the mismatched thymine (underlined in the previous sequence). One of the exonucleases RecJ, ExoVII, or ExoI then degrades the site before DNA polymerase resynthesizes the gap in the strand.[5]
Base excision repair
[edit]AP site formation is a common occurrence in dsDNA. It is the result of spontaneous hydrolysis and the activity of DNA glycosylases as an intermediary step in base excision repair. These AP sites are removed by AP endonucleases, which effect single strand breaks around the site.[5]
Nucleotide excision repair
[edit]Nucleotide excision repair, not to be confused with base excision repair, involves the removal and replacement of damaged nucleotides. Instances of crosslinking, adducts, and lesions (generated by ultraviolet light or reactive oxygen species) can trigger this repair pathway. Short stretches of single stranded DNA containing such damaged nucleotide are removed from duplex DNA by separate endonucleases effecting nicks upstream and downstream of the damage. Deletions or mutations which affect these nucleases instigate increased sensitivity to ultraviolet damage and carcinogenesis. Such abnormalities can even impinge neural development.
In bacteria, both cuts executed by the UvrB-UvrC complex. In budding yeast, Rad2 and the Rad1-Rad10 complex make the 5' and 3' cuts, respectively. In mammals, the homologs XPG and XPF-ERCC1 affect the same respective nicks.[9]
Double-strand break repair
[edit]Double-strand breaks, both intentional and unintentional, regularly occur in cells. Unintentional breaks are commonly generated by ionizing radiation, various exogenous and endogenous chemical agents, and halted replication forks. Intentional breaks are generated as intermediaries in meiosis and V(D)J recombination, which are primarily repaired through homologous recombination and non-homologous end joining. Both cases require the ends in double strand breaks be processed by nucleases before repair can take place. One such nuclease is Mre11 complexed with Rad50. Mutations of Mre11 can precipitate ataxia-telangiectasia-like disorder.[9]
V(D)J recombination involves opening stem-loops structures associated with double-strand breaks and subsequently joining both ends. The Artemis-DNAPKcs complex participates in this reaction. Although Artemis exhibits 5' → 3' ssDNA exonuclease activity when alone, its complexing with DNA-PKcs allows for endonucleasic processing of the stem-loops. Defects of either protein confers severe immunodeficiency.[9]
Homologous recombination, on the other hand, involves two homologous DNA duplexes connected by D-loops or Holliday junctions. In bacteria, endonucleases like RuvC resolve Holliday junctions into two separate dsDNAs by cleaving the junctions at two symmetrical sites near the junction centre. In eukaryotes, FEN1, XPF-ERCC1, and MUS81 cleave the D-loops, and Cce1/Ydc2 processes Holliday junctions in mitochondria.[9]
Meganucleases
[edit]The frequency at which a particular nuclease will cut a given DNA molecule depends on the complexity of the DNA and the length of the nuclease's recognition sequence; due to the statistical likelihood of finding the bases in a particular order by chance, a longer recognition sequence will result in less frequent digestion. For example, a given four-base sequence (corresponding to the recognition site for a hypothetical nuclease) would be predicted to occur every 256 base pairs on average (where 4^4=256), but any given six-base sequence would be expected to occur once every 4,096 base pairs on average (4^6=4096).
One unique family of nucleases is the meganucleases, which are characterized by having larger, and therefore less common, recognition sequences consisting of 12 to 40 base pairs. These nucleases are particularly useful for genetic engineering and Genome engineering applications in complex organisms such as plants and mammals, where typically larger genomes (numbering in the billions of base pairs) would result in frequent and deleterious site-specific digestion using traditional nucleases.
See also
[edit]- HindIII
- Ligase
- Micrococcal nuclease
- Nuclease protection assay
- P1 nuclease
- PIN domain
- Polymerase
- Serratia marcescens nuclease (benzonase)
- S1 nuclease
References
[edit]- ^ Nishino T, Morikawa K (December 2002). "Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors". Oncogene. 21 (58): 9022–32. doi:10.1038/sj.onc.1206135. PMID 12483517.
- ^ a b Rittié L, Perbal B (June 2008). "Enzymes used in molecular biology: a useful guide". Journal of Cell Communication and Signaling. 2 (1–2): 25–45. doi:10.1007/s12079-008-0026-2. PMC 2570007. PMID 18766469.
- ^ Linn S, Arber W (April 1968). "Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form". Proceedings of the National Academy of Sciences of the United States of America. 59 (4): 1300–6. Bibcode:1968PNAS...59.1300L. doi:10.1073/pnas.59.4.1300. PMC 224867. PMID 4870862.
- ^ Arber W, Linn S (1969). "DNA modification and restriction". Annual Review of Biochemistry. 38: 467–500. doi:10.1146/annurev.bi.38.070169.002343. PMID 4897066.
- ^ a b c d e f Nishino T, Morikawa K (December 2002). "Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors". Oncogene. 21 (58): 9022–32. doi:10.1038/sj.onc.1206135. PMID 12483517.
- ^ Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, et al. (May 1993). "The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments". The EMBO Journal. 12 (5): 1781–95. doi:10.2210/pdb4rve/pdb. PMC 413397. PMID 8491171.
- ^ a b c d Nishino T, Morikawa K (December 2002). "Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors". Oncogene. 21 (58): 9022–32. doi:10.1038/sj.onc.1206135. PMID 12483517.
- ^ Nishino T, Morikawa K (December 2002). "Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors". Oncogene. 21 (58): 9022–32. doi:10.1038/sj.onc.1206135. PMID 12483517.
- ^ a b c d Nishino T, Morikawa K (December 2002). "Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors". Oncogene. 21 (58): 9022–32. doi:10.1038/sj.onc.1206135. PMID 12483517.
