User talk:Evolution and evolvability/Archive 2015
Do not edit this page. This is the archive of User talk:Evolution and evolvability for the year 2015. (Please direct any additional comments to the current talk page.) See the annual archives for 2015, 2016, 2017, 2018, 2019, 2020. |
Welcome!
Hi, Evolution and evolvability. Welcome to Wikipedia! Thank you for your contributions. I hope you like the place and decide to stay. Our intro page contains a lot of helpful material for new users—please check it out! If you need help, visit Wikipedia:Questions, ask me on my talk page, or place {{helpme}} on this page, followed by your question, and someone will show up shortly to answer your questions. jonkerz ♠talk 15:59, 19 October 2013 (UTC)
A barnstar for you!

|
The Original Barnstar |
| Thanks for your excellent article on the PA clan and many other contributions to Protein and RNA families! Alexbateman (talk) 11:15, 15 January 2014 (UTC) |
- @Alexbateman: I just realised that I never thanked you for this. I'm glad that the PA clan article and Protein superfamily are useful. Hopefully more of the well-defined superfamilies and folds will get pages over time. T.Shafee(Evo﹠Evo)talk 11:07, 11 June 2015 (UTC)
If you get bored of enzymes, I have a suggestion...
The gene article still contains this aesthetically unpleasant low-res image File:Rna-codons-protein.png that some idiot edited and uploaded ages ago :) Opabinia regalis (talk) 09:53, 6 March 2015 (UTC)
- @Opabinia regalis: Ha, thanks for pointing me in its direction. I'll definitely have a stab at it, and maybe a couple of other images in at now that I've had a look. Being part of the Enzyme FAR has made me more comfortable with the idea of editing bigger articles. T.Shafee(Evo﹠Evo)talk 10:31, 6 March 2015 (UTC)
- Thanks! That image is annoying because it's my only remaining one that hasn't been moved to commons, and now it's stranded because it's so old the NIH doesn't use it anymore, and the fact that it's still hosted on their server somewhere is good enough evidence for PD here but not on commons.
- I don't have it on hand right now but there's evidence somewhere around here that the bigger/broader wikipedia articles are systematically crappier. So I say go for as big an article as you have the patience for :) Opabinia regalis (talk) 20:21, 7 March 2015 (UTC)
Image lifecycle
Hey, I really like your question about image lifecycle. Nobody has looked into image halflife. I would agree with your conclusion: images are there for a very long time regardless of content. I have seen typos on images that have persisted for years and there are a lot of unclear and unrelated images in articles —I have shied away from deleting many. Unfortunately I don't have the space and time to download and parse the wikimedia commons dump right now —next week?. My guess is like your, namely they are pretty immutable; but the distribution may be surprising as quality would influence half-life. Although, I should do that in a dozen years as your images will probably smash all records given their quality! Matteo--Squidonius (talk) 21:59, 9 March 2015 (UTC)
Idea for glycolysis
Just a thought - your glycolysis diagram is much superior to the images already in the glycolysis article (File:Glycolysis2.svg and especially File:Glycolysis.jpg), but doesn't show the names of the enzymes associated with each step. If you numbered the arrows, as usually suggested on commons for text labels - and maybe colored the arrows for steps that consume/produce ATP? - your image would be a great substitute for the existing ones in the article, which get the job done but are very busy and cluttered-looking. Opabinia regalis (talk) 04:39, 23 March 2015 (UTC)
- @Opabinia regalis: Thanks! I know what you mean, an article as important as glycolysis probably needs a little better than File:Glycolysis.jpg. I was thinking of doing a second version with more information. I like your coloured arrows idea for ATP production/use. May have to do something for NADH and H2O in reactions too. Not sure yet how to get enzyme and metabolite names in without cluttering but it should be ok. What do you reckon? T.Shafee(Evo﹠Evo)talk 11:08, 23 March 2015 (UTC)
- Of course I can't think of an example now, but IIRC commons often recommends using numbers instead of text labels so that the image can be reused in non-English projects. I don't know that enzyme names change that much, but numbers would be easier to fit into the image. Highlighting the NADH and H2O in the reactions is a good idea. Thinking out loud, I'm imagining a second image containing the (color-coded?) enzyme names, presented in the article in a frameless table so that it looks like one continuous image. But that probably sounds too complicated; there's a reason I'm not a graphic designer :) Opabinia regalis (talk) 04:49, 24 March 2015 (UTC)
- That sounds great. When it's easy, the usual thing to do is to produce a version with English labels and a separate file with numbers. In a well-ordered .svg file, it's often possible for knowledgeable people (i.e., not me) to change the labels to whatever language they want.
- If it sounds like fun, there's also an image map system at Commons. When you hover, you get more (text) information. I don't know much about it, but I could probably find someone who did. Just ping me if you want to know more about that. WhatamIdoing (talk) 16:57, 25 March 2015 (UTC)
- @WhatamIdoing:I was thinking of using wikilinkable image annotations (something like this). The main thing that I don't like is how when you then open the full-sized image the text isn't there. T.Shafee(Evo﹠Evo)talk 03:04, 26 March 2015 (UTC)
- Do you think that commons:Help:Gadget-ImageAnnotator would do what you want? WhatamIdoing (talk) 00:24, 27 March 2015 (UTC)
- @WhatamIdoing:I was thinking of using wikilinkable image annotations (something like this). The main thing that I don't like is how when you then open the full-sized image the text isn't there. T.Shafee(Evo﹠Evo)talk 03:04, 26 March 2015 (UTC)
- Of course I can't think of an example now, but IIRC commons often recommends using numbers instead of text labels so that the image can be reused in non-English projects. I don't know that enzyme names change that much, but numbers would be easier to fit into the image. Highlighting the NADH and H2O in the reactions is a good idea. Thinking out loud, I'm imagining a second image containing the (color-coded?) enzyme names, presented in the article in a frameless table so that it looks like one continuous image. But that probably sounds too complicated; there's a reason I'm not a graphic designer :) Opabinia regalis (talk) 04:49, 24 March 2015 (UTC)
@Opabinia regalis, WhatamIdoing, and Seppi333: Thank you for all your help so far. Here is my first attempt at using image annotations: Template:Glycolysis summary. I've transcluded it into the Glycolysis article. Any thoughts? T.Shafee(Evo﹠Evo)talk 12:49, 27 March 2015 (UTC)
- Wow, this is great, nice work!! The annotation template is a really good idea; I didn't even know it existed. I have one ridiculously picky comment - it might just be my browser, but the 'split' in the bottom arrow (which is a really clever idea) appears in the full view to be solid white, not transparent. Opabinia regalis (talk) 06:30, 28 March 2015 (UTC)
- @Opabinia regalis: Hah, good point! I always forget to check waht I've made transparent, and what I've made white! The main down-side of both image annotator and image map is that neither works properly with the mobile phone interface! T.Shafee(Evo﹠Evo)talk 11:04, 28 March 2015 (UTC)
- That looks great. Thanks for doing that! WhatamIdoing (talk) 15:28, 28 March 2015 (UTC)
- @Opabinia regalis: Hah, good point! I always forget to check waht I've made transparent, and what I've made white! The main down-side of both image annotator and image map is that neither works properly with the mobile phone interface! T.Shafee(Evo﹠Evo)talk 11:04, 28 March 2015 (UTC)
- I normally think the mobile interface is embarrassingly terrible, but I just checked this on my phone and the image and annotations display OK to me. The only problem is that the image is too wide for the vertical orientation, but it displays as expected horizontally.
- Is there such a thing as a featured template? :) Opabinia regalis (talk) 19:50, 28 March 2015 (UTC)
Hi there, I'm pleased to inform you that I've begun reviewing the article Gene you nominated for GA-status according to the criteria. ![]() This process may take up to 7 days. Feel free to contact me with any questions or comments you might have during this period. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 01:40, 10 April 2015 (UTC)
This process may take up to 7 days. Feel free to contact me with any questions or comments you might have during this period. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 01:40, 10 April 2015 (UTC)
The article Gene you nominated as a good article has been placed on hold ![]() . The article is close to meeting the good article criteria, but there are some minor changes or clarifications needing to be addressed. If these are fixed within 7 days, the article will pass; otherwise it may fail. See Talk:Gene for things which need to be addressed. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 17:20, 11 April 2015 (UTC)
. The article is close to meeting the good article criteria, but there are some minor changes or clarifications needing to be addressed. If these are fixed within 7 days, the article will pass; otherwise it may fail. See Talk:Gene for things which need to be addressed. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 17:20, 11 April 2015 (UTC)
The article Gene you nominated as a good article has failed ![]() ; see Talk:Gene for reasons why the nomination failed. If or when these points have been taken care of, you may apply for a new nomination of the article. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 03:41, 19 April 2015 (UTC)
; see Talk:Gene for reasons why the nomination failed. If or when these points have been taken care of, you may apply for a new nomination of the article. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 03:41, 19 April 2015 (UTC)
- @Cerebellum: Thanks for acting as reviewer. You're definitely right that the citations are below par. Will try to address that and resubmit soon. T.Shafee(Evo﹠Evo)talk 04:09, 19 April 2015 (UTC)
- @Evolution and evolvability: Great, let me know when you resubmit and I'll be happy to review again. --Cerebellum (talk) 11:29, 20 April 2015 (UTC)
Interview for The Signpost
This is being sent to you as a member of WikiProject Molecular and Cellular Biology The WikiProject Report would like to focus on WikiProject Molecular and Cellular Biology for a Signpost article. This is an excellent opportunity to draw attention to your efforts and attract new members to the project. Would you be willing to participate in an interview? If so, here are the questions for the interview. Just add your response below each question and feel free to skip any questions that you don't feel comfortable answering. Multiple editors will have an opportunity to respond to the interview questions, so be sure to sign your answers. If you know anyone else who would like to participate in the interview, please share this with them. Thanks, Rcsprinter123 (express) @ 16:23, 9 May 2015 (UTC)
Have been meaning to do this for weeks...

|
The Graphic Designer's Barnstar | |
| Awarded to Evolution and evolvability for contributing many outstanding images to Wikipedia, including this highly informative and unique overview of gene structure. Adrian J. Hunter(talk•contribs) 04:24, 10 June 2015 (UTC) |
- Seconded! It's really nice to see wiki articles illustrated with professional and modern-looking images, and those annotated templates are great. Opabinia regalis (talk) 21:09, 10 June 2015 (UTC)
- @Adrian J. Hunter and Opabinia regalis: Thank you both! I've really enjoyed making some updated images for wikipedia and it's good to know that they're clear to others as well! T.Shafee(Evo﹠Evo)talk 11:07, 11 June 2015 (UTC)
Thank you
Thank you for thanking me for the edits to the "glycolysis" article. Cruithne9 (talk) 05:20, 20 July 2015 (UTC)
The article Gene you nominated as a good article has passed ![]() ; see Talk:Gene for comments about the article. Well done! If the article has not already been on the main page as an "In the news" or "Did you know" item, you can nominate it to appear in Did you know. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 17:41, 25 July 2015 (UTC)
; see Talk:Gene for comments about the article. Well done! If the article has not already been on the main page as an "In the news" or "Did you know" item, you can nominate it to appear in Did you know. Message delivered by Legobot, on behalf of Cerebellum -- Cerebellum (talk) 17:41, 25 July 2015 (UTC)
- Congratulations! :) Opabinia regalis (talk) 18:09, 25 July 2015 (UTC)
Your GA nomination of Catalytic triad
Hi there, I'm pleased to inform you that I've begun reviewing the article Catalytic triad you nominated for GA-status according to the criteria. ![]() This process may take up to 7 days. Feel free to contact me with any questions or comments you might have during this period. Message delivered by Legobot, on behalf of Stigmatella aurantiaca -- Stigmatella aurantiaca (talk) 22:40, 10 August 2015 (UTC)
This process may take up to 7 days. Feel free to contact me with any questions or comments you might have during this period. Message delivered by Legobot, on behalf of Stigmatella aurantiaca -- Stigmatella aurantiaca (talk) 22:40, 10 August 2015 (UTC)
Your GA nomination of Catalytic triad
The article Catalytic triad you nominated as a good article has passed ![]() ; see Talk:Catalytic triad for comments about the article. Well done! If the article has not already been on the main page as an "In the news" or "Did you know" item, you can nominate it to appear in Did you know. Message delivered by Legobot, on behalf of Stigmatella aurantiaca -- Stigmatella aurantiaca (talk) 16:01, 19 August 2015 (UTC)
; see Talk:Catalytic triad for comments about the article. Well done! If the article has not already been on the main page as an "In the news" or "Did you know" item, you can nominate it to appear in Did you know. Message delivered by Legobot, on behalf of Stigmatella aurantiaca -- Stigmatella aurantiaca (talk) 16:01, 19 August 2015 (UTC)
- Congratulations again! Really excellent work on this article. You could put some biochemistry on the main page now for DYK.... :) Opabinia regalis (talk) 07:53, 21 August 2015 (UTC)
Catalytic triad
Hi,
Thanks for the great work on the Catalytic triad article. I have asked Neil Rawlings (down the corridor) to comment on the article, but he is a bit busy at the moment. But if you can wait a few weeks I'm sure he'll give you some great feedback. Alexbateman (talk) 11:02, 24 August 2015 (UTC)
- @Alexbateman: That's absolutely fine. There's no rush, and I'm always particularly happy to get input from people who don't typically wikipede. T.Shafee(Evo﹠Evo)talk 12:00, 24 August 2015 (UTC)
Half-life of protein backbone
I see that you have given the half-life of a protein backbone as being 500 years in the article Proteolysis, I wonder if you have a source for it. I'm just slightly puzzle because I have seen widely different figures given for peptide bonds, from 7 years here to over a hundred years here. I would assume the high figure of 500 has something to do with the exposure to water (or rather the lack thereof) in the folded interior of a protein, but I'm not sure because parts of most proteins would be exposed to solvent. Hzh (talk) 00:41, 1 September 2015 (UTC)
- @Hzh: Thank you for reminding me. I'd meant to go back and pull out the reference for it:
:Radzicka, Anna; Wolfenden, Richard (January 1996). "Rates of Uncatalyzed Peptide Bond Hydrolysis in Neutral Solution and the Transition State Affinities of Proteases". Journal of the American Chemical Society. 118 (26): 6105–6109. doi:10.1021/ja954077c.
- It's actually for dipeptides, so the effect of solvent exclusion for a folded protein is ignored. The other references you have could be interesting for comparison. T.Shafee(Evo﹠Evo)talk 00:57, 1 September 2015 (UTC)
- Many thanks. I would assume the side-chain group has a strong effect on the rate of hydrolysis, maybe bulky groups side chains are less stable than gly-gly. The book might explain it more but only parts of it can be read. Hzh (talk) 01:09, 1 September 2015 (UTC)
WP:INTREF
There are a bajillion links that go to WP:INTREF, which is now a double-redirect (along with the links to the other pages). I think we should keep Help:Introduction to referencing/1 as-is, then have Help:Introduction to referencing (VE)/1. That OK? Otherwise thank you very much for creating the VE section, it was much needed. FYI you can transclude the verifiability and reliable sources sections in the VE pages, since they will be the same. — MusikAnimal talk 16:47, 13 November 2015 (UTC)
- I've bold done both. So we'll keep Help:Introduction to referencing/1, etc, for the wikimarkup, just because that's the long-standing home and there's a lot of cleanup involved to get the redirects right. This is a highly visible page (link in the protection reasons, etc), so double-redirects are no good =P I also refactored the reliable sources to Help:Introduction to referencing/Reliable sources. It is now sourced in the VE pages. Thanks again for making this happen. Cheers — MusikAnimal talk 17:49, 13 November 2015 (UTC)
- @MusikAnimal: Thanks for helping out! I'd not appreciated how much or a problem the double-redirects were. Is it possible to just change WP:INTREF to redirect to the ...(WM) version? I guess it's not the biggest deal in the world, but I feel the consistency could be useful. Good idea with transcluding to ensure that the info remains synchronised! I'm a big fan of trying to make sure that pages can be more easily maintained (hence the
{{intro to}}template). If you've the time and inclination, it'd be great if you could have a quick look over the new Help:Introduction to referencing (VE) pages. T.Shafee(Evo﹠Evo)talk 09:23, 15 November 2015 (UTC)- Looks great! I guess I did not take the time to check how many redirects we are working with, and turns out there aren't many. Sorry about that, we can go back to ...(WM) naming scheme. I can handle that and the redirects, and will do so shortly.But the big question remains... which referencing guide do we link to as the default? E.g. {{refimprove}} along with similar templates are atop millions of articles. Changing the "citations" link to the new VE pages will have a profound effect, I think. And honestly, in most cases it's SO much easier with VisualEditor, so I'm in favour of sending our users there, but we should seek broader consensus first. I can open up a discussion at WP:VPP, how does that sound?Another thing, I have concerns Book:Wikipedia: The Missing Manual might be a little too much information for the users we're targeting on Help pages. How about linking to Help:Wiki markup instead? It's fairly comprehensive and not a long read. — MusikAnimal talk 19:29, 15 November 2015 (UTC)
- @MusikAnimal: All very good points. Thanks for handling the behind-the-scenes redirects. For default referencing recommendations, I agree that it's reached a point where a WP:VPP is appropriate, and making sure that the relevant groups that might be interested are directed towards it. I've no experience in these matters, so I'm happy for you to put the proposal up! My plan for the two base links is one for switching between WM/VE editing methods and two get to the fullest information repository. At the moment, I envisage that the WP:VisualEditor/User guide link will be at the base of all intro_to...(VE) pages. So similarly, I think it'd be good to have a consistent, equivalent link to put at the bottom of all intro_to...(WM) pages (including talk pages, tables, images etc). Now that I look at it, H:MARKUP does actually cover all that so is probably the closest equivalent of the WP:VE/UG. I suspect most readers will take the tutorials as self-contained modules, with the minimum necessary instruction to achieve basic editing proficiency. T.Shafee(Evo﹠Evo)talk 02:05, 16 November 2015 (UTC)
- Ah, I realized we can't change all the links to the VE guide just yet. Anonymous users still don't have access to VisualEditor! Such a shame. It's only a matter of time before this happens, and when it does we'll revisit the proposal. For now we'll stick with what we have.Did you say you were okay with changing the "full manual" link to H:MARKUP? The other issue with the book is that it goes well beyond wikimarkup, also documenting general Wikipedia best practices. I also worry if it is outdated, as much of the Book namespace seems to be. The idea behind Help pages should be to keep it really simple and brief, so I think H:MARKUP is fitting in this case. I have a long term goal of dumbing down all the other help pages, such as Wikipedia:Your first article. Out of all of the help pages I think WP:INTREF is the best example of how we should approach them. Brevity, easy navigation, illustrations, friendly wording, etc. — MusikAnimal talk 02:49, 16 November 2015 (UTC)
- @MusikAnimal: All very good points. Thanks for handling the behind-the-scenes redirects. For default referencing recommendations, I agree that it's reached a point where a WP:VPP is appropriate, and making sure that the relevant groups that might be interested are directed towards it. I've no experience in these matters, so I'm happy for you to put the proposal up! My plan for the two base links is one for switching between WM/VE editing methods and two get to the fullest information repository. At the moment, I envisage that the WP:VisualEditor/User guide link will be at the base of all intro_to...(VE) pages. So similarly, I think it'd be good to have a consistent, equivalent link to put at the bottom of all intro_to...(WM) pages (including talk pages, tables, images etc). Now that I look at it, H:MARKUP does actually cover all that so is probably the closest equivalent of the WP:VE/UG. I suspect most readers will take the tutorials as self-contained modules, with the minimum necessary instruction to achieve basic editing proficiency. T.Shafee(Evo﹠Evo)talk 02:05, 16 November 2015 (UTC)
- Looks great! I guess I did not take the time to check how many redirects we are working with, and turns out there aren't many. Sorry about that, we can go back to ...(WM) naming scheme. I can handle that and the redirects, and will do so shortly.But the big question remains... which referencing guide do we link to as the default? E.g. {{refimprove}} along with similar templates are atop millions of articles. Changing the "citations" link to the new VE pages will have a profound effect, I think. And honestly, in most cases it's SO much easier with VisualEditor, so I'm in favour of sending our users there, but we should seek broader consensus first. I can open up a discussion at WP:VPP, how does that sound?Another thing, I have concerns Book:Wikipedia: The Missing Manual might be a little too much information for the users we're targeting on Help pages. How about linking to Help:Wiki markup instead? It's fairly comprehensive and not a long read. — MusikAnimal talk 19:29, 15 November 2015 (UTC)
- @MusikAnimal: Thanks for helping out! I'd not appreciated how much or a problem the double-redirects were. Is it possible to just change WP:INTREF to redirect to the ...(WM) version? I guess it's not the biggest deal in the world, but I feel the consistency could be useful. Good idea with transcluding to ensure that the info remains synchronised! I'm a big fan of trying to make sure that pages can be more easily maintained (hence the
{{Intro to}}
Thanks for all the work you've done on this template and the related pages. One puzzle, for me: the template appears to force all paragraphs to be fully justified. That's not at all the norm for Wikipedia help pages. How would you feel about changing the alignment to left justification, which is the norm? -- John Broughton (♫♫) 21:49, 25 November 2015 (UTC)
- @John Broughton:You're right that it's not the norm. I've not got particularly strong views.Justified text looks slightly neater to me, but then I can imagine it more easily misformatting on narrow screens. T.Shafee(Evo﹠Evo)talk 22:22, 25 November 2015 (UTC)
- My personal philosophy with regard to readers is avoid surprises. Fully justified text is surprising because it's not used elsewhere. (I also personally don't like it - I find the odd spacing of words to be off-putting - but that's just a personal opinion.)
- Since you don't have particularly strong views on this, and I do, would you mind if I changed the justification? (If you do mind, then the next step is to get some other opinions, of course.) -- John Broughton (♫♫) 17:44, 26 November 2015 (UTC)
- @John Broughton:Changing the template's text alignment is fine by me. It can always be turned back if there's a sudden swell of support for a justified format! T.Shafee(Evo﹠Evo)talk 08:52, 27 November 2015 (UTC)
- Thanks. -- John Broughton (♫♫) 19:55, 27 November 2015 (UTC)
Maybe
If you're interested in getting access to review articles for biomedical topics, then you might look at this new opportunity: Wikipedia:Annual Reviews. The usual rules seem to apply: you need to have made 500+ edits, to have been around for six months or more, and not have access to this particular source through work or school. Assuming you don't already have access elsewhere, then you easily meet the requirements. WhatamIdoing (talk) 17:32, 16 December 2015 (UTC)
More FP
So, which of your protein illustrations do you like best? Catalytic triad? I feel like we should nominate a couple at FP to set the standard for what a good, professional-quality protein image looks like, because crappy renders from 2006 sure aren't it. Opabinia regalis (talk) 00:00, 18 December 2015 (UTC)
- Hi, I had a quick look through my archive for just a 'standard' ribbon cartoon as an illustrative example and realised that I'm almost always doing something extra. My favourites are probably:
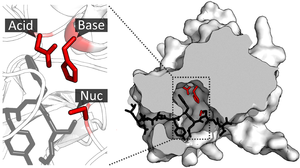
- File:Catalytic triad of TEV protease.png - how a detail can be highlighted and how cutaways can be useful
- File:Galactosidase enzyme.png - a 'standard' ribbon diagram to show a structure with various details highlighted in place
- File:Hexokinase induced fit.png - a nice combination of schematic and surface to make a single point
- File:Cut 1UVB.png - another useful cut-away, works as a pair with File:Uncut 1UVB.png
- File:Serpin mechanism (S to R).png - finally, I'm pretty happy with how clearly this one shows the serpin motion
I'm open to suggestion, of course, if you have opinions as to which. As always, I'm very happy to know that my images are useful! I might also do a bit of an update on the MCB diagram guide (although it's not exactly high-traffic). T.Shafee(Evo﹠Evo)talk 11:54, 19 December 2015 (UTC)
- Ooh, I like all of these! I think the catalytic triad or the serpin would be my picks in terms of 'simple enough to explain to laypeople but complex enough to look impressive' :) Seems that FPC has lots of self-noms - do you want to do the honors or should I?
- More updates on the diagram guide would be great - maybe it will get more traffic if there's more meat on the bones there. Opabinia regalis (talk) 05:35, 21 December 2015 (UTC)
- Also, is File:Galactosidase enzyme.png galactosidase (in the filename) or glucosidase (in the caption and PDB description) or does that enzyme do both? Opabinia regalis (talk) 05:41, 21 December 2015 (UTC)
- I would love you to nominate them! Somehow I still feel awkward self-nominating, even though there's little functional difference! Well spotted that File:Galactosidase enzyme.png is misnamed. I think I was planning on doing a different enzyme and changed half-way through the upload. I'll rename it on commons! T.Shafee(Evo﹠Evo)talk 05:50, 21 December 2015 (UTC)
- I just took a look at the FP criteria and it appears there are Rules about image pixel size which these fall slightly short of. (So does the DNA clamp, but, well, 2006.) Any chance of re-rendering the tried at a larger size? (Apparently 1500px is the preferred minimum.)
- Sorry, one other minor point: now that I've looked at the serpin on my phone, it's hard to distinguish the dark gray and light gray/white. That phone is old and crappy and has a cracked screen and is getting replaced soon, so take that for what it's worth :) Opabinia regalis (talk) 20:26, 21 December 2015 (UTC)
- I'll fix the resolutions tonight. Should be easy enough. I'll also increase the tone difference between ther serpin and its target protease. You're right that it's actually too subtle when I use an outlined render. The ideal is an image that is as easily interpretable as possible. T.Shafee(Evo﹠Evo)talk 00:54, 22 December 2015 (UTC)
- I would love you to nominate them! Somehow I still feel awkward self-nominating, even though there's little functional difference! Well spotted that File:Galactosidase enzyme.png is misnamed. I think I was planning on doing a different enzyme and changed half-way through the upload. I'll rename it on commons! T.Shafee(Evo﹠Evo)talk 05:50, 21 December 2015 (UTC)
Ok, I've updated all the images to be at least 1500px wide. Also, File:Galactosidase enzyme.png → File:Glucosidase enzyme.png renaming complete. I actually took the opportunity to save an original vector version to the commons (File:Hexokinase induced fit.svg). I might actually go though enzyme and replace the images with .svg versions, since vectors are pretty good for web browsers. Either way, I think the images on the bullet-point list are good to go. T.Shafee(Evo﹠Evo)talk 10:19, 22 December 2015 (UTC)
- Great, thanks! I am off to the airport for
cattle callholiday travel and may not get to this till after Christmas, but on the other hand if plane wifi doesn't suck for once, I might do it tonight :) Happy holidays in either case! Opabinia regalis (talk) 00:30, 23 December 2015 (UTC)- Done, finally. Hope you had a good Christmas! Opabinia regalis (talk) 21:46, 26 December 2015 (UTC)
- Thanks Opabinia regalis! I'm still getting used to the Australian Summer-Christmas. Hope you've had a good holiday too, despite the travel! T.Shafee(Evo﹠Evo)talk 01:21, 27 December 2015 (UTC)
- Done, finally. Hope you had a good Christmas! Opabinia regalis (talk) 21:46, 26 December 2015 (UTC)
Evolutionary algorithms
Hi, I just wonder what makes you worrying about the content of my userpage (is it appearing in some lists?), spend your time on Wikipedia at all? Btw, I'm currently studying NEAT algorithms. Neuroevolution of augmenting topologies — Preceding unsigned comment added by Ilyadorosh (talk) 11:26, 27 December 2015 (UTC)
- @Ilyadorosh: Hi the level of duplicated content just made me a little uncomfortable. I realise you weren't trying to impersonate me, but the page ended up being misleading (e.g. listing my images and barnstars as yours and giving my email address as your contact details). I look forward to your contributions to the evolutionary algorithm articles! T.Shafee(Evo﹠Evo)talk 03:44, 5 January 2016 (UTC)