User:Kinkreet/Protein Science/Enzyme mechanisms
The rate of reaction for the uncatalysed glycine decarboxylation have a half-time of 1.1 billion years, whereas that for peptide cis-trans isomerization have a half-time of 23 seconds. Reactions needed to sustain life needs to occur at a much faster pace (20ms half-time), and so biological enzymes must be present.
Chemical catalysis is not enough, as they can usually only improve the rates by 10-104 fold, whereas biological enzymes can improve typically by 106 to 1012 fold, some even up to 1017 folds. Most biological enzymes are specific and produce no byproduct, and is able to function at milder conditions not possible with chemical catalysis; also, it can be regulated tightly.
The study of enzyme mechanism is a cycle of creating a 3D model (for the whole protein as well as the active site), generate a possible reaction mechanism and testing it using kinetic studies, to see how well the model fits the data. The model is then refined to fit the data better.
In disease states, certain enzymes are upregulated and so they can be a marker for disease; for example, when intracellular bacteria invades a cell, autophagy proteins are upregulated. These, however, is hard to detect quickly. When cells are damaged, the cell debris which includes enzymes are released into the bloodstream. There are organ/tissue-specific enzymes which can be used as indicators of where the damage has occurred.
| Enzyme | Marker for |
|---|---|
| Acid phosphatase | Prostatic carcinoma |
| Alanine aminotransferase | Hepatocellular damage |
| Alkaline phosphatase | Cholestatic liver disease/Osteoblast activity in bone disease |
| Amylase | Cell damage in acute pancreatitis |
| Aspartate aminotransferase | Hepatocellular damage, muscle damage (e.g. myocardiac infarction) |
| Creatine kinase | Muscle damage and acute myocardiac infarction |
| γ-glutamyl transpeptidase | Liver cell damage |
| Lactate dehydrogenase | Muscle damage |
Michaelis-Menten
[edit]
The general equation for an enzyme substrate reaction can be written as follows:
E + S ↔ ES ↔ EP ↔ E + P
First, the enzyme and substrate must bind to form an enzyme-substrate complex, then the substrate is converted to the product(s), which is subsequently released from the enzyme.
Michaelis-Menten kinetics is a simplified model of the above reaction, and relates reaction rate, v, with substrate concentration [S]. It was developed by German biochemist Leonor Michaelis and Canadian physician Maud Menten, on their study on invertase.
where , and denote the rate constants. To make this simplification acceptable, we must assume that once the product is formed, it dissociates so quickly that virtually no reverse reaction (EP → ES) is observed. Because of this, can also be taken as , the turnover number or the maximum number of substrate molecules converted to product per enzyme molecule per second.
If we assume that the concentration of the enzyme-substrate complex does not change over the course of the reaction (quasi-steady-state assumption), i.e. .
Divide by :
where
By multiplying by , we get
Here, represents the maximum rate achieved by the system, at maximum (saturating) substrate concentrations. The Michaelis constant is the substrate concentration at which the reaction rate is half of , and thus is a rough approximation to the inverse indication of binding affinity. Biochemical reactions involving a single substrate are often assumed to follow Michaelis–Menten kinetics, without regard to the model's underlying assumptions.
As is indicative of binding affinity, and is an indicator for reaction rate, the two can be combined as to represent the overall efficiency of an enzyme. When ES → P is not rate limiting, the rate of the reaction is determined mainly by , which is limited by diffusional collision rates. Therefore, is limited 108 – 1010 /M.s because the number of binding events between substrate and enzyme is limited by diffusion. Such enzymes are termed to be catalytic perfect; enzymes such as catalase, fumarase and acetylcholinesterase are enzymes close to achieving catalytic perfection. Most of these enzymes have such high efficiency because of tunnelling, where there are amino acids next to the active site which guides the substrate to the active site, and so every substrate that encounters the enzyme is catalysed.
Isoenzymes are enzymes which catalyse the same reaction but have different amino acids in their active sites and thus have different catalytic efficiency. This is useful in an industrial setting to select the most efficient isoenzyme to use. Also, an enzyme will have preferences for different substrates, for example chymotrypsin have a preference for bulky and aromatic side chains on the carbonyl group of the bond (peptide or ester bonds) to be cleaved.
As [S]→0,
As [S]→∞,
Assumptions made:
- E + P → ES is negligible
- quasi-steady-state is achieved, and thus cannot be used to estimate parameters at the beginning or end of a reaction, where there is a lack of ES and S, respectively.
- Assume [S] >> [ES] so that , meaning that ES → E + P is the rate-limiting reaction.
- If we assume [S] << KM, then is a measure of an enzyme's catalytic efficiency.
Plots
[edit]Plotting reaction rate verse substrate concentration will not allow you to get an accurate estimation of vmax. Other plots must be used, and each have their own advantages and disadvantages.
Lineweaver-Burke plot
[edit]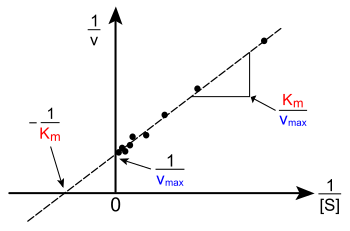
The Lineweaver-Burke plot takes the reciprocal of both sides of the Michaelis-Menten equation.
Taking the reciprocal gives
The Lineweaver-Burke plot have the major disadvantage of being inaccurate. This is because it gives undue weight to small [S], which in the plot have high values; therefore, small errors in v will lead to a large error when reciprocated. This is a major source of error because it is at these low concentrations of substrate where the errors are the most significant. Likewise, the large [S] are all crowded towards the left and is hard to use it to determine a straight line. These are given insufficient weight although they tend to be the most accurate. For these reasons, it is not usually used to determine kinetic parameters.
It is however useful for determining the type of enzyme inhibition (competitive, uncompetitve, or mixed)
Hofstee-Eadie plot
[edit]It can be derived from the Michaelis–Menten equation as follows:
invert and multiply with :
Rearrange:
Isolate v:
The Hofstee-Eadie plot have an advantage over the Lineweaver-Burke plot because it gives more equal weighting for different [S] values, because [S] is plotted with v, and higher [S], higher v will be also. However, a large error may still remain because both axis is dependent on v.
Hanes-Woolf plot
[edit]The equation can be derived from the Michaelis–Menten equation as follows:
invert and multiply by [S]:
Rearrange:
This also gives a more even weighting of data, and tend to be the most accurate.
Acetaldehyge dehydrogenase
[edit]Ethanol in the body is oxidized using alcohol dehydrogenase to acetaldehyde
- CH3CH2OH + NAD+ → CH3CHO + NADH + H+
Acetaldehyde is then oxidized by acetaldehyde dehydrogenase:

Most people have two forms of acetaldehyde dehydrogenase: Aldehyde dehydrogenase 2 (encoded by the ALDH2 gene) functions in the mitochondria and has a low ; and one in the cytosol (encoded by ALDH1) has a high <math<K_M</math>. Some persons of far-Eastern descent have a single point mutation (G → A) at exon 12 of the ALDH2 gene which translates to a causes a dominant single amino acid mutation (Q487K) in their mitochondrial acetaldehyde dehydrogenase gene, resulting in the ALDH2K enzyme.[1] This mutation increases the the mitochondrial dehydrogenase's affinity for NAD+, and thus less likely for it to carry out the reaction through to the product, thus it is less active. The majority of the oxidation must then be carried using the cytosolic form, which is much slower.
Acetaldehyde is more toxic than ethanol, and is the cause of hangovers and acetaldehyde poisoning, including the characteristic flushing of the skin and increased heart and respiration rates. Thus people with the ALDH2K mutant blushes even after a small amount of alcohol is consumed, also called Asian flush.
Inhibition
[edit]Many types of drugs alter enzyme actions by activating/inactivating/promoting/inhibiting the enzyme. Inhibitors are molecules that bind reversibly to enzymes and decrease their activty; inactivators are inhibitors that binds irreversibly.
| Drug | Target |
|---|---|
| Aspirin | Prostaglandin synthase |
| Penicillin | Glycopeptide transpeptidase |
| Methotrexate | Dihydrofolate reductase |
| AZT | HIV reverse transcriptase |
| Ritonavir | HIV protease |
| Viagra | cGMP phosphodiesterase |
Competitive inhibition
[edit]Competitive inhibition is where the inhibitor competes with the substrate for the active site, because the inhibitor and substrate have similar shapes. Once bound to the active site, it does not get catalyzed and thus will remain there until it dissociates; meanwhile, the substrate cannot bind, and thus reaction rate is reduced. Methotrexate have a similar structure to dihydrofolate, and thus can competitively bind dihydrofolate reductase.
Some bacteria need to generate folic acid in order to survive and grow. Folic acid acts as cofactors in the synthesis of purine, pyrimidine and other amino acids; it is synthesized using dihydrofolate synthetase. para-aminobenzoic acid (PABA) is a structural part of folic acid, and is incorporated during synthesis. Sulfanilamide is a analog of PABA and thus is used as an antibiotic to competitively inhibit dihydrofolate synthetase; the bacteria cannot grow and will die. Because humans takes in folic acid, it is not toxic at the doses given in the antibiotics.
References
[edit]- ^ Crabb, David; Xiao, Qing (1998). "Studies on the Enzymology of Aldehyde Dehydrogenase-2 in Genetically Modified HeLa Cells". Alcoholism: Clinical and Experimental Research. 22 (4): 780. doi:10.1111/j.1530-0277.1998.tb03867.x.






![{\displaystyle k_{1}[E][S]=k_{-1}[ES]+k_{cat}[ES]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/4d6dee8ade169b92820e4a561b2bb959bca83586)
![{\displaystyle k_{1}([E]_{0}-[ES])[S]=[ES](k_{-1}+k_{cat})}](https://wikimedia.org/api/rest_v1/media/math/render/svg/0e870e16b382dcb28b6dc26fee51a0ffa2955f84)
![{\displaystyle [S]k_{1}[E]_{0}-[S]k_{1}[ES]=[ES](k_{-1}+k_{cat})}](https://wikimedia.org/api/rest_v1/media/math/render/svg/fd3e5cda5c0c8606fefe54f0eb4a855cfe2a1320)
![{\displaystyle [S]k_{1}[E]_{0}=[ES](k_{-1}+k_{cat})+[S]k_{1}[ES]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/49bffe5c128f96b640dad7114817196ef23e7340)
![{\displaystyle [S]k_{1}[E]_{0}=[ES](k_{-1}+k_{cat}+[S]k_{1})}](https://wikimedia.org/api/rest_v1/media/math/render/svg/cc2bd3892c1d2b57ea50cae4a25291fd857f3570)
![{\displaystyle {\frac {[S]k_{1}[E]_{0}}{(k_{-1}+k_{cat}+[S]k_{1})}}=[ES]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/f58e0b12fca7319f0eb5acabc578b4639429123a)
![{\displaystyle {\frac {[S][E]_{0}}{({\frac {k_{-1}+k_{cat}}{k_{1}}}+[S])}}=[ES]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/5d8b99a17f3e12e93e064b225257e6b83dab7d03)
![{\displaystyle [ES]={\frac {[E]_{0}[S]}{K_{m}+[S]}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/3a3a565bba720ec33d9ebdbaaaeeacdeed812d71)

![{\displaystyle v={\frac {d[P]}{dt}}=V_{\max }{\frac {[S]}{K_{m}+[S]}}=k_{\text{cat}}[E]_{0}{\frac {[S]}{K_{m}+[S]}}.}](https://wikimedia.org/api/rest_v1/media/math/render/svg/e37fa824ed43fdf208e14849dcb20d2f55235c25)




![{\displaystyle v_{0}\approx {\frac {v_{max}[S]}{K_{M}}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/a6c26cb2019a2634a6ca146d43e34840545e082f)


![{\displaystyle V={\frac {V_{\max }[S]}{K_{m}+[S]}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/8fa5b35e10661c7592ae5bc59429f1033a4acbf8)
![{\displaystyle {1 \over V}={{K_{m}+[S]} \over V_{\max }[S]}={K_{m} \over V_{\max }}{1 \over [S]}+{1 \over V_{\max }}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/262e1440a8ad30a692b153178eabbf6e7f45d48f)
![{\displaystyle v={{V_{\max[}S]} \over {K_{m}+[S]}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/ffe8e1abf595d0f2b2941797aa98e1784cb21452)
![{\displaystyle {V_{\max } \over v}={{V_{\max(}K_{m}+[S])} \over {V_{\max[}S]}}={{K_{m}+[S]} \over {[S]}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/78b5e6e7562d64f81b548ce47817ffe1215d26af)
![{\displaystyle V_{\max }={{{vK_{m}} \over {[S]}}+{{v[S]} \over {[S]}}}={{vK_{m}} \over {[S]}}+v}](https://wikimedia.org/api/rest_v1/media/math/render/svg/ce05f9ad1eb9473b9c34da535fa9aa939236e1d9)
![{\displaystyle v=-K_{m}{v \over {[S]}}+V_{\max }}](https://wikimedia.org/api/rest_v1/media/math/render/svg/2f636c0144f921be4d81f8de9d867ea9e0f3deb5)
![{\displaystyle {[S] \over v}={{[S](K_{m}+[S])} \over {V_{\max[}S]}}={{K_{m}+[S]} \over {V_{\max }}}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/205ac16a4c5734f25049d68170b38505062b7342)
![{\displaystyle {[S] \over v}={1 \over V_{\max }}[S]+{K_{m} \over V_{\max }}}](https://wikimedia.org/api/rest_v1/media/math/render/svg/3bb781047fcb26179df1b93d6d8c0cde115a59b4)
