User:Kharris439/sandbox
o-Succinylbenzoate synthase
[edit]o-Succinylbenzoate synthase (OSBS) (EC 4.2.1.113)[1] is an enzyme encoded by the menC gene in E.coli, and catalyzes the dehydration of 2-succinyl-6-hydroxy-2,4-
| o-succinylbenzoate synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 The conformation of o-succinylbenzoate synthase when it is bound to the Mg cation and its product, OSB, is illustrated here. The cyan-colored R groups represent the R groups that interact with the Mg cation. The red- and cyan-colored R groups represent the R groups that interact with OSB. The single yellow-colored R group represents Lys 133, which has been shown to act as both the catalytic base and acid in the dehydration reaction. | |||||||||
| Identifiers | |||||||||
| EC no. | 4.2.1.113 | ||||||||
| CAS no. | 97089-83-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
cyclohexadiene-1-carboxylate (SHCHC) to form 4-(2'-carboxyphenyl)-4-oxobutyrate, also called o-succinylbenzoate or OSB, hence the name of the enzyme.[1][2][3][4][5] This reaction is the fourth step in the menaquinone biosynthetic pathway, which is used by bacteria to synthesize menaquinone, also known as vitamin K2.[6]
Classification
[edit]OSBS belongs to the muconate lactonizing enzyme subgroup of the enolase superfamily. The systematic name of this enzyme is (1R,6R)-6-hydroxy-2-succinylcyclohexa-2,4-diene-1-carboxylate hydrolyase (2-succinylbenzoate-forming). Other common names include: o-succinylbenzoic acid synthase and OSB synthase.
Menaquinone biosynthetic pathway
[edit]The menaquinone biosynthetic pathway consists of nine enzymatic reactions, ultimately resulting in the synthesis of vitamin K. This pathway is quite similar in both plants and bacteria, however the final product of the pathway varies slightly in the two organisms. The final product in plants and some cyanobacteria is phylloquinone, which functions as an electron transporter in photosynthesis. The final product in bacteria and archaea is menaquinone, which is involved in anaerobic respiration.[6] The structures of the two end-products are illustrated below.

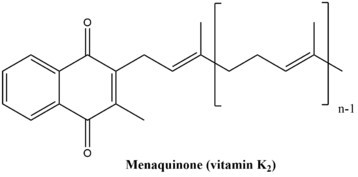
Phylloquinone is commonly called "vitamin K1." Menaquinone is commonly called "vitamin K2." Both fall under the common name "vitamin K." This pathway is not present in humans and other animals, however vitamin K is essential for blood coagulation and therefore must be consumed. Good sources of vitamin K include vegetables (kale, spinach, broccoli, Brussels sprouts, cauliflower, etc.), and fish, liver, meat, and eggs (though these contain smaller amounts of vitamin K than do vegetables.)[7]
Structure
[edit]OSBS is a monomeric protein consisting of two domains: the "capping domain" and the "barrel domain," both of which are characteristic of the enolase superfamily. The enzyme's active site is located at the interface of the two domains, with the acid-base chemistry occurring on the barrel domain.[8] OSBS requires the presence of Mg2+ to function. Mg2+ stabilizes the intermediate during the reaction.[1]
Activity
[edit]The reaction catalyzed by OSBS is shown below:

The reaction involves the dehydration of SHCHC to form OSB. OSBS has one substrate, SHCHC, and two products, water and OSB.
Homologues in other organisms
[edit]The binding of OSB to OSBS's active site consists mainly of indirect interactions via water molecules or hydrophobic interactions. This lack of strict specificity and catalysis could possibly simplify the evolution of the shape and volume of the active site, meaning that OSBS could serve as a starting point for the evolution of new enzymes with new functions in the enolase superfamily. These homologues could catalyze completely different reactions, but because they maintain an active site similar to that of OSBS, the substrate and intermediate of the new reaction would be structurally similar to that of OSBS.[3][4][5]
One such homologue has already been identified: OSBS from Amycolatopsis. OSBS from Amycolatopsis was first identified as N-acylamino acid racemase (NAAAR) because it was found to catalyze the racemization of N-acylamino acids. However, this ability was discovered for commercial reasons, and considering racemization of N-acylamino acids does not occur in Amycolatopsis, its actual job in the bacteria itself was unknown. In 1999, it was discovered that NAAAR's protein sequence was quite similar to another protein with an unknown function in Bacillus subtilis. Both proteins were found to efficiently catalyze the same reaction as OSBS in E.coli, and so this was deemed their "correct" function. "NAAAR" was properly renamed to OSBS. The hypothesis stated in the first paragraph of this section helps explain how the OSBS from Amycolatopsis can also catalyze the racemization of N-acylamino acids, as well as the diversity of catalytic differences among the enzymes of the enolase superfamily.[3][4][5]
References
[edit]- ^ a b Klenchin, Vadim A.; Taylor Ringia, Erika A.; Gerlt, John A.; Rayment, Ivan (2003). "Evolution of Enzymatic Activity in the Enolase Superfamily: Structural and Mutagenic Studies of the Mechanism of the Reaction Catalyzed by o-Succinylbenzoate Synthase from Escherichia coli" (PDF). Biochemistry, 42, 14427-14433.
- ^ Sharma, V.; Meganathan, R.; Hudspeth, M.E.S (August 1993). "Menaquinone (vitamin K2) biosynthesis: cloning, nucleotide sequence, and expression of the menC gene from Escherichia coli". Journal of Bacteriology.
- ^ a b c Thompson, Thomas B.; Garrett, James B.; Taylor, Erika A.; Meganathan, R.; Gerlt, John A.; Rayment, Ivan (2000). "Evolution of Enzymatic Activity in the Enolase Superfamily: Structure of o-Succinylbenzoate Synthase from Escherichia coli in Complex with Mg2+ and o-Succinylbenzoate". Biochemistry, 39, 10662-10676.
- ^ a b c Palmer, David R. J.; Garrett, James B.; Sharma, V.; Meganathan, R.; Babbitt, Patricia C.; Gerlt, John A. (1999). "Unexpected Divergence of Enzyme Function and Sequence: "N-Acylamino Acid Racemase" is o-Succinylbenzoate Synthase". Biochemistry, 38, 4252-4258.
- ^ a b c Taylor Ringia, Erika A.; Garrett, James B.; Thoden, James B.; Holden, Hazel M.; Rayment, Ivan; Gerlt, John A. (2004). "Evolution of Enzymatic Activity in the Enolase Superfamily: Functional Studies of the Promiscuous o-Succinylbenzoate Synthase from Amycolatopsis". Biochemistry, 43, 224-229.
- ^ a b van Oostende, C; Widhalm, JR; Furt, F; Ducluzeau, AL; Basset, GJC (2011). "Phylloquinone (Vitamin K1): function, enzymes and genes in Advances in Botanical Research". eds Fabrice Rébeillé and Roland Douce, 59: 229-61, Academic Press (Amsterdam).
- ^ "Vitamin K". University of Maryland Medical Center. 06/21/2011.
{{cite web}}: Check date values in:|date=(help) - ^ "Research Interests". Gerlt Lab - Molecular and Cellular Biology - University of Illinois.

