User:Dhoenngg/sandbox
Archaeol
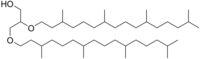
[edit]Archaeol is one of the main core membrane lipids of archaea, existing ubiquitously in archaea cells. It’s broadly used as a biomarker for ancient archaea, especially methanogens, activity.
| |
| Names | |
|---|---|
| IUPAC name
2,3-Bis(3,7,11,15-tetramethylhexadecoxy)propan-1-ol
| |
| Other names
Archaeol lipid; 2,3-Di-O-phytanyl-sn-glycerol; 2,3-Bis[(3,7,11,15-tetramethylhexadecyl)oxy]-1-propanol
| |
| Identifiers | |
3D model (JSmol)
|
|
| MeSH | archaeol+lipid |
PubChem CID
|
|
| |
| Properties | |
| C43H88O3 | |
| Molar mass | 653.174 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Chemistry
[edit]
Five kinds of archaeol have been identified so far, contributing to the complexity of the membrane lipids. The most typical form is 2,3-di-O-phytanyl-sn-glycerol, with two phytanyl chains binding to the position of 2 and 3 of glycerol by ether bonds. This is different from bacteria and eukarya that use 1,2-sn-glycerol[1]. Natural archaeol has 3R, 7R, 11R configurations for the three chiral centers in the isoprenoid chains. The two phytanyl chains can form a 36-member ring to yield macrocyclic archaeol. Hydroxylated archaeol has phytanyl chains hydroxylated at the first methine group, while sesterterpanyl archaeol have the phytanyl side chains with C25 sesterterpanyl chains, substituting at C2 of glycerol or at both carbons. Unsaturated archaeol, with the same carbon skeleton but one or multiple double bonds in the phytanyl side chains is also discovered[2].
Archaeal liposomes typically demonstrate extremely low permeability for molecules and ions, even including protons. The ion permeability induced by ionophores are also quite low, and only comparable to that of egg phosphatidylcholine at 37 ˚C when the temperature rises up to c.a. 70˚C[3][4].




Biological role and synthesis
[edit]Archaeol has been found in all analyzed archaea so far, at least trace amount. It represents 100% of the diether core lipids in most neutrophilic halophiles[2] and sulfur-dependent thermophiles (though their most core lipids are tetraether lipids). Methanogens contain hydroxyarchaeol and macrocyclic other than the standard archaeol, and sesterterpanyl-chain-containing archaeol is characteristic of alkaliphilic extreme halophiles. It’s noteworthy that tetraether lipids are also widely present in archaea[1].
Compared to bacteria and eukarya, the isoprenoid side chains of archaeol are highly branched. This structural difference is believed to provide archaea membrane with low permeability over the whole growth temperature range to help archaea better adapt to extreme environments[5].
Ether membrane lipids have also been discovered in some aerobic and anaerobic bacteria, including lipids with one ester bond and one ether bond to alkyl chains. Many strictly anoxic bacteria and a few aerobic species also contain plasmalogens (Pla), which has a alkyl chain bound to sn-1 position of the glycerol via a vinyl-ether bond. Similar to archaea, these lipids are thought to increase the resistivity of bacteria to adverse environments. More stunning is the discovery of nonisoprenoid dialkyl glycerol diether lipids(DGD) and branched dialkyl glycerol tetraether lipids (brGDGT). These lipids are only different from archaea ether lipids in the side chains and binding positions on the glycerol. DGD is reported in thermophilic bacteria, a few mesophilic bacteria and aggregating myxobacteria[6][7].
Archaeol is usually in the form of phospholipid in archaea cells. The synthetic pathway of fully-saturated archaeol phospholipid includes sequentially the synthesis of isoprenoid side chains, ether linkage to glycerol-1-phosphate backbone, CDP archaeol formation, polar head group attachment and saturation of double bonds. . Tetraether lipids may be synthesized afterwards by dimerization reaction via a head-to-head linkage[8].
Archaea feature different synthesis pathway of isoprenoid chains from either bacteria or eukarya. The building blocks for isoprenoid are universally C5 units: isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). Typically, the two compounds are synthesized in bacteria via 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate pathway (MEP/DOXP pathway), and mevalonate (MVA) pathway in most eukarya. The synthesis of the two compounds in archaea follows an alternate MVA pathway which differs from the classic MVA pathway in the last three steps and shares the remaining four steps[8].
Used as a lipid biomarker
[edit]Archaeol in the sediments typically comes from the hydrolysis of membrane lipids during diagenesis. Due to its well preservation, it is often detected and used by organic geochemists in its original form as a biomarker for archaea activity, especially for methanogen biomass and activity. It is used by Michinari Sunamura et al. to directly measure the methanogens in the sediments of Tokyo Bay[9], and also used by Katie L. H. Lim et al. as an indicator of methanogenesis in water-saturated soils[10]. C. A. McCartney et al. used it as a proxy for methane production in cattle[11].
It was used as a biomarker by Richard D. Pancost et al. in order to reconstruct the Holocene biogeochemistry in ombrotrophic peatlands[12]. A pilot study led by Ian D. Bull et al. also used archaeol as a biomarker to reveal the differences between fermenting digestive systems in foregut and hindgut of ancient herbivorous mammals[13]. Additionally, the ratio of archaeol to caldarchaeol was proposed as a salinity proxy in highland lakes, providing a tool for paleosalinity studies[14]. The authors speculated that it is because of different degradation kinetics of intact archaeol and caldarchaeol.
Measurement
[edit]Bligh-Dyer process is extensively used in the extraction of core lipids, usually followed by fractionation or derivatization. Similar procedures are proposed by Kazuhiro Demizu et al.[15] and Sadami Ohtsubo et al.[16] involving acid Bligh and Dyer extraction[17], acid treatment, methanolysis and fractionation, with the core lipids finally being subjected to chromatography.
For determination of archaeol concentration, chromatography technologies are widely used, including high-performance liquid chromatography(HPLC)[15][16][18], gas chromatography(GC)[19], and supercritical fluid chromatography(SFC)[20][21], with mass spectrometry(MS) often applied to aid the analysis.
See also
[edit]References
[edit]- ^ a b Koga, Y Nishihara, M Morii, H Akagawa-Matsushita, M. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. OCLC 680443863.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b "Archaeal lipids and their biotechnological applications". World Journal of Microbiology and Biotechnology. 11: 115–131. 1995.
- ^ Yamauchi, Kiyoshi; Doi, Kuniyuki; Kinoshita, Masayoshi; Kii, Fumiko; Fukuda, Hideki (October 1992). "Archaebacterial lipid models: highly salt-tolerant membranes from 1,2-diphytanylglycero-3-phosphocholine". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1110 (2): 171–177. doi:10.1016/0005-2736(92)90355-p. ISSN 0005-2736.
- ^ Yamauchi, Kiyoshi; Doi, Kumiyuki; Yoshida, Yoichi; Kinoshita, Masayoshi (March 1993). "Archaebacterial lipids: highly proton-impermeable membranes from 1,2-diphytanyl-sn-glycero-3-phosphocoline". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1146 (2): 178–182. doi:10.1016/0005-2736(93)90353-2. ISSN 0005-2736.
- ^ Koga, Yosuke (2012). "Thermal Adaptation of the Archaeal and Bacterial Lipid Membranes". Archaea. 2012: 1–6. doi:10.1155/2012/789652. ISSN 1472-3646. PMC 3426160. PMID 22927779.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Grossi, Vincent; Mollex, Damien; Vinçon-Laugier, Arnauld; Hakil, Florence; Pacton, Muriel; Cravo-Laureau, Cristiana (2015). "Mono- and Dialkyl Glycerol Ether Lipids in Anaerobic Bacteria: Biosynthetic Insights from the Mesophilic Sulfate Reducer Desulfatibacillum alkenivorans PF2803T". Applied and Environmental Microbiology. 81 (9): 3157–3168. doi:10.1128/AEM.03794-14.
- ^ Lorenzen, Wolfram; Ahrendt, Tilman; Bozhüyük, Kenan A J; Bode, Helge B (2014-05-11). "A multifunctional enzyme is involved in bacterial ether lipid biosynthesis". Nature Chemical Biology. 10 (6): 425–427. doi:10.1038/nchembio.1526. ISSN 1552-4450.
- ^ a b Jain, Samta (2014). "Biosynthesis of archaeal membrane ether lipids". Frontiers in Microbiology. 5. doi:10.3389/fmicb.2014.00641. PMC 4244643. PMID 25505460.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Sunamura, Michinari; Koga, Yosuke; Ohwada, Kouichi (1999-11-01). "Biomass Measurement of Methanogens in the Sediments of Tokyo Bay Using Archaeol Lipids". Marine Biotechnology. 1 (6): 562–568. doi:10.1007/PL00011811. ISSN 1436-2228.
- ^ Lim, Katie L. H.; Pancost, Richard D.; Hornibrook, Edward R. C.; Maxfield, Peter J.; Evershed, Richard P. (2012). "Archaeol: An Indicator of Methanogenesis in Water-Saturated Soils". Archaea. 2012: 1–9. doi:10.1155/2012/896727. ISSN 1472-3646. PMC 3512251. PMID 23226972.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Dewhurst, R. J.; Yan, T.; Bull, I. D.; McCartney, C. A. (2013-02-01). "Assessment of archaeol as a molecular proxy for methane production in cattle". Journal of Dairy Science. 96 (2): 1211–1217. doi:10.3168/jds.2012-6042. ISSN 0022-0302. PMID 23261373.
- ^ Pancost, Richard D.; McClymont, Erin L.; Bingham, Elizabeth M.; Roberts, Zoë; Charman, Dan J.; Hornibrook, Edward R.C.; Blundell, Anthony; Chambers, Frank M.; Lim, Katie L.H. (November 2011). "Archaeol as a methanogen biomarker in ombrotrophic bogs". Organic Geochemistry. 42 (10): 1279–1287. doi:10.1016/j.orggeochem.2011.07.003.
- ^ Gill, Fiona L.; Dewhurst, Richard J.; Dungait, Jennifer A.J.; Evershed, Richard P.; Ives, Luke; Li, Cheng-Sen; Pancost, Richard D.; Sullivan, Martin; Bera, Subir (May 2010). "Archaeol – a biomarker for foregut fermentation in modern and ancient herbivorous mammals?". Organic Geochemistry. 41 (5): 467–472. doi:10.1016/j.orggeochem.2010.02.001.
- ^ Wang, Huanye; Liu, Weiguo; Zhang, Chuanlun L.; Jiang, Hongchen; Dong, Hailiang; Lu, Hongxuan; Wang, Jinxiang (January 2013). "Assessing the ratio of archaeol to caldarchaeol as a salinity proxy in highland lakes on the northeastern Qinghai–Tibetan Plateau". Organic Geochemistry. 54: 69–77. doi:10.1016/j.orggeochem.2012.09.011.
- ^ a b Demizu, Kazuhiro; Ohtsubo, Sadami; Kohno, Shuhei; Miura, Isao; Nishihara, Masateru; Koga, Yosuke (1992). "Quantitative determination of methanogenic cells based on analysis of ether-linked glycerolipids by high-performance liquid chromatography". Journal of Fermentation and Bioengineering. 73 (2): 135–139. doi:10.1016/0922-338x(92)90553-7. ISSN 0922-338X.
- ^ a b Ohtsubo, S (May 1993). "A sensitive method for quantification of aceticlastic methanogens and estimation of total methanogenic cells in natural environments based on an analysis of ether-linked glycerolipids". FEMS Microbiology Ecology. 12 (1): 39–50. doi:10.1016/0168-6496(93)90023-z. ISSN 0168-6496.
- ^ Bligh, E. G.; Dyer, W. J. (August 1959). "A RAPID METHOD OF TOTAL LIPID EXTRACTION AND PURIFICATION". Canadian Journal of Biochemistry and Physiology. 37 (8): 911–917. doi:10.1139/o59-099. ISSN 0576-5544.
- ^ Martz, Robert F.; Sebacher, Daniel I.; White, David C. (February 1983). "Biomass measurement of methane forming bacteria in environmental samples". Journal of Microbiological Methods. 1 (1): 53–61. doi:10.1016/0167-7012(83)90007-6. ISSN 0167-7012.
- ^ Smith, G.C.; Floodgate, G.D. (October 1992). "A chemical method for estimating methanogenic biomass". Continental Shelf Research. 12 (10): 1187–1196. doi:10.1016/0278-4343(92)90078-x. ISSN 0278-4343.
- ^ Holzer, Gunther U.; Kelly, Patrick J.; Jones, William J. (July 1988). "Analysis of lipids from a hydrothermal vent methanogen and associated vent sediment by supercritical fluid chromatography". Journal of Microbiological Methods. 8 (3): 161–173. doi:10.1016/0167-7012(88)90017-6. ISSN 0167-7012.
- ^ King, Jerry (2002-01-22), "Supercritical Fluid Technology for Lipid Extraction, Fractionation, and Reactions", Lipid Biotechnology, CRC Press, ISBN 9780824706197, retrieved 2019-05-31
