User:Azackta1/sandbox
Group Contribution towards Eukaryotic DNA replication Article
[edit]Chosen references
[edit]Balancing eukaryotic replication asymmetry with replication fidelity
- Coordinated replication of eukaryotic nuclear genomes is asymmetric, with copying of a leading strand template preceding discontinuous copying of the lagging strand template
- Replication is catalyzed by DNA polymerases α, δ and ɛ
- Recent studies suggest that Pol ɛ is normally the primary leading strand replicase, whereas most synthesis by Pol δ occurs during lagging strand replication
- One strand of DNA is synthesized continuously, but the other is made as Okazaki fragments that are later joined together
- After initial priming by pol α, pol δ takes over the synthesis on the lagging strand.
- pol δ can correct errors made by pol α, thereby protecting genome stability
- Replication protein A (RPA) assembles on the ssDNA immediately after the helicases unwind double-stranded DNA,creating a replication fork.
DNA Replication and Checkpoint Control in S Phase
- Good overview of DNA replication during the S phase. Has nice figures but not sure if they are available for PD use.
Initation of DNA replication: functional and evolutionary aspects
- Great overview of the pre-RC formation and I like that similar to the above "Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity" article this journal article is more recent.
- This article also looks at aspects that are conserved and similar across all eukarytotes regarding replication initiation.
Eukaryotic DNA replication: from pre-replication complex to initiation complex
- Pre-RC explanation and overview.
- Really good overview of the process of protien kinase role in initiation of replication.
- Reference to MCM as a replicative helicase, combined with the MCM article above, this is a great reference to back up the MCM information.
Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases
- Good scientific article for referencing MCM function.
- This article gives a bit more information to reference Cdc45 as a likely cofactor in the replicative helicase.
DNA Replication in Eukaryotic Cells
- brings together results from different organisms to provide a coherent model of the events of initiation and discusses:
- The Origin Recognition Complex
- The Cdc6 Protein
- The Cdt1 Protein
- The Mcm2–7 Protein Complex
- Assembly and function of the pre-replicative complex
- Transition to replication
- Assembly of the DNA polymerases
- control by kinases
- Intra-S-phase checkpoint
The DNA replication fork in eukaryotic cells
- Cellular replication fork proteins
- DNA Polymerase α/Primase Complex
- Replication Protein A (RPA)
- Replication Factor C (RFC)
- Proliferating Cell Nuclear Antigen (PCNA)
- DNA Polymerases δ and ɛ
- FEN1 and RNaseHI
- DNA Helicases / DNA2 Helicase
- Mini-chromosome maintenance (MCM) proteins
- Mechanisms of synthesis at replication fork
- Primosome Assembly
- Polymerase Switching
- Maturation of Okazaki Fragments
- Replication Fork proteins and Cell Cycle
- S-Phase Checkpoint Control
- PCNA-p21 Interaction
- Regulation of Telomere Length
Initiation of Eukaryotic DNA Replication: Regulation and Mechanisms
- The Eukaryotic Cell Cycle
- Factors Required for the Initiation of DNA Replication
- The Organization of Replication-Initiation Factors on Chromatin
- Cell Cycle Control by Checkpoints
Regulation of DNA replication licensing
- DNA replication is tightly regulated to occur only once per cell cycle
- discusses how DNA licensing is a mechanism to guarantee this aim
Overview of the Cell Cycle and Its Control
- Discusses the process of chromosome replication during the S phase of the cell cycle
Loading and activation of DNA replicative helicases: the key step of initiation of DNA replication
- To duplicate chromosomal DNA, doublestranded DNA must first be unwound by helicase, which is loaded to replication origins and activated during the DNA replication initiation step.
- Discusses the common features of, and differences in, replicative helicases between prokaryotes and eukaryotes.
Regulation of Eukaryotic DNA Replication
- Origins of Replication
- Initiation Molecules and Mechanisms
- Cell-Cycle Control of DNA Replication
Genome-wide model for the normal eukaryotic DNA replication fork
- Discusses lagging strand replication
- Pol δ is primarily a lagging strand polymerase during replication across the entire nuclear genome
Possible Images
[edit]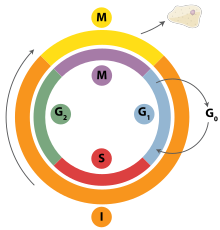



Preliminary Outline for Eukaryotic DNA replication Article
[edit]Lead Section:
- Add more information other than "DNA replication in eukaryotes is much more complicated than in prokaryotes, although there :are many similar aspects". Include the location of DNA replication, but also include an introduction to the different :proteins that are used to form the replication complexes.
Location in the Cell:
- Discuss where eukaryotic DNA replication takes place and why. Discuss why eukaryotic chromosome are replicated only once per :cell cycle (Watson p.239).
- replication only occurs during the S phase of the cell cycle
- during this phase, all of the DNA chromosome needs to be replicated exactly once, incomplete replication causes :inappropriate links between daughter chromosomes.
Multiple Origins of Replication:
- Discuss the need for multiple origins of replication.
- origins are separated by 30kb
- multiple origins are needed to ensure that the whole chromosome is replicated during the S phase.
- Discuss how origins of replication are activated and inactivated and why.
Mechanisms of Replication:
- Origins and Initiation:
- Discuss the formation of the prereplicative complex
- assembly of the pre-RC is an ordered process
- initiated by the association of the origin recognition complex (ORC) with the replicator
- ORC recruits additional proteins, Cdc6 and Cdt1
- These proteins function together to assemble the eukaryotic DNA helicase, the Mcm2-7 complex
- Completes the formation of the pre-RC
- Discuss how the formation of the pre-RC also regulates a single round of DNA replication during each cell cycle
- Discuss how the activation of the pre-RC leads to the assembly of the eukaryotic replication fork
- As cells enter into the S phase of the cell cycle, Cdk and Ddk phosphorylate replication proteins to trigger the initiation of replication
- Activation of these kinases leads to the release of Cdc6 and Cdt1
- Events that lead to DNA unwinding are poorly understood, but are thought to require the activity of the Mcm complex and recruitment of auxiliary factors and DNA Pol δ and Pol ε
- DNA Pol α/primase is recruited after Pol δ and Pol ε, once at the site of origin, Pol α/primase synthesizes an RNA primer and briefly extends it
- the resulting primer:template junction is recognized by the eukaryotic sliding clamp loader (RF-C), which assembles the sliding clamp (PCNA) at these sites
- Pol ε recognizes this primer and begins leading-strand synthesis
- After a period of DNA unwinding, Pol α/primase synthesizes additional primers, which allow the initiation of lagging strand DNA synthesis by DNA Pol δ
- Replication Fork
- Formation of replication fork and different polymerases used in pro- vs eukaryotic cells
- Leading and Lagging Strand information
- Other important factors such as DNA gyrase, ss binding proteins (SSB) and sliding clamp/loaders.
- All referenced in Watson, etal. pgs 239-250 very well
- Regulation:
- Location in the cell cycle plays vital role with various checkpoints
- Protein, CDK, and cyclin factors all help control progression through checkpoints
- Termination:
- Telomere regions and telomerase
Five Pillars of Wikipedia
[edit]Wikipedia operates on five fundamental principles that can be summarized as the Five Pillars of Wikipedia.
- Wikipedia is an encyclopedia:
- Wikipedia is best represented as a collaboration of encyclopedias, almanacs, and gazettes. Wikipedia is not intended for personal use as in advertising, personal articles or essays, or personal blogs. Wikipedia is also not a dictionary,newspaper, or a source for lists of links to other online sources.
- Wikipedia is created from a neutral point of view:
- Wikipedia only submits articles that explain topics from a broad point of view without siding one way or another. All articles must be verifiability accurate and include references.
- Wikipedia includes content that is free for anyone to edit:
- Wikipedia articles are not owned by one person so content within Wikipedia is free to edit, modify, or add information to a page. You must follow copyright laws and not plagiarize from sources.
- Wikipedia must remain friendly between users:
- Wikipedia asks that you remain friendly and in good faith while interacting with other Wikipedians. Never start an editing war, retain proper etiquette, and Wikipedia likes to remind you that there are 4,155,593 articles [1] on the English Wikipedia to work on and discuss.
- Wikipedia does not have firm rules:
- Wikipedia rules are not final and are subject to change. Wikipedia encourages its users to be bold and to make corrections and additions to articles.
Summary of characteristics of target article
[edit]For our Molecular Biology course, we will be editing an article from a stub class to somewhere between a B or GA class article. Listed is a summary of the two types of article classes we will be working towards achieving this semester.
B class article:
- is complete without any major issues
- contains enough information to satisfy a casual reader but not for someone who is needing more information for research
- is complete but lacks style and may need to be edited by a senior editor
- needs to comply with the Manual of Style and style guidelines
- needs more work to reach good article standards[2][3]
GA class article:
- is useful for all readers, contain factually accurate and verifiable information
- has no obvious problems, are very well written
- are broad in coverage, neutral in point of view, stable and have illustrations
- editing by subject and style editors can be helpful or compare to a similar article for useful differences
- meets good article standards[2][4]
PubMed Articles
[edit]An article from Retrovirology: Cell type and specific requirements for thiol/disulfide exchange during HIV-1 entry and infection, focuses its studies on the role of disulfide bonds in HIV-1 infection to further the understanding of the characterization of the process. The authors use established cell lines and/or CXCR4-tropic laboratory-adapted virus for previous findings and extended their focus to CCR5, primary human macrophages, and CD4+T lymphocytes. The authors found that the nonspecific thiol/disulfide exchange inhibitor, 5-5'-dithiobis(2-nitrobenzoic acid) (DTNB), significantly reduced HIV-1 entry and infection in cell lines, human monocyte-derived macrophages, and PHA-stimulated peripheral blood mononuclear cells (PBMC).[5] The authors findings were able to demonstrate the role of thiol/disulfide exchange in HIV-1 entry in primary T lymphocytes and monocyte-derived macrophages. The authors also concluded the utilization of PDI could be relevant to the HIV-1 entry and establishment of the virus reservoirs in resting CD4+ cells.[5]
An article from PNAS: Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry, focuses its studies on a pseudo type ebolavirus glycoprotein and it's mode of entry into a cell. The authors recently showed that binding of the receptor binding region (RBR) of the ebolavirus glycoprotein (GP) and infection by the GP psuedotyped virons increase during cell adhesion independently of mRNA or protein synthesis.[6] The authors expanded on their previous findings and provided evidence for a proposed model of infection: during cell adhesion, there is a RBR binding partner that translocates intracellular vesicles from the virus to the cell surface.[6] The authors used a suspended 293T cell line that contained RBR binding sites to test their proposed model. The results of the experiment caused the authors to reexamine an initial claim that lymphocytes contain a pool of RBR binding partner and concluded that the mode of entry is by membrane-trafficking that translocates an RBR binding partner to the cell surface in which the Ebola virus utilizes for virus entry.
References
[edit]- ^ http://en.wikipedia.org/wiki/Wikipedia:FIVEPILLARS
- ^ a b http://en.wikipedia.org/wiki/Wikipedia:Quality#Grades
- ^ http://en.wikipedia.org/wiki/Wikipedia:Version_1.0_Editorial_Team/Assessment/B-Class_criteria
- ^ http://en.wikipedia.org/wiki/Wikipedia:GA
- ^ a b Stantchev TS, Paciga M, Lankford CR, Schwartzkopff F, Broder CC, Clouse KA (2012). "Cell-type specific requirements for thiol/disulfide exchange during HIV-1 entry and infection". Retrovirology. 9: 97. doi:10.1186/1742-4690-9-97. PMC 3526565. PMID 23206338.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b Dube D, Schornberg KL, Shoemaker CJ; et al. (September 2010). "Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry". Proc. Natl. Acad. Sci. U.S.A. 107 (38): 16637–42. doi:10.1073/pnas.1008509107. PMC 2944755. PMID 20817853.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: date and year (link) CS1 maint: multiple names: authors list (link)
