Tryptophan 7-halogenase
| Tryptophan 7-Halogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of Trp 7-halogenase PrnA from PDB 2AQJ | |||||||||
| Identifiers | |||||||||
| EC no. | 1.14.19.9 | ||||||||
| CAS no. | 198575-11-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Tryptophan 7-halogenase (EC 1.14.19.9, PrnA, RebH) is an enzyme with systematic name L-tryptophan:FADH2 oxidoreductase (7-halogenating).[1][2] This enzyme catalyses the following chemical reaction:
- tryptophan + FADH2 + Cl− + O2 + H+ 7-chloro-L-tryptophan + FAD + 2 H2O
The enzyme can use bromide ions (Br−) in place of chloride (Cl−).[3]
Background
[edit]Tryptophan 7-halogenase is a member of a class of enzymes known as flavin-dependent halogenases. Prior to tryptophan 7-halogenase's discovery, it was thought that all halogen atoms in metabolites were incorporated by the action of haloperoxidases, another class of halogenases dependent on metal centers such as vanadium or heme, or perhydrolases, a class of halogenases that generates peracids that in turn oxidize halide ions to hypohalous acids, which act as halogenating agents.[4] These enzymes halogenate without substrate specificity and regioselectivity.[5] The first tryptophan 7-halogenase was isolated in 1995 by Dairi et al. after a comparison in amino acid sequence revealed no similarity with previously known haloperoxidases.[6] In contrast with haloperoxidases, flavin-dependent halogenases are much narrower in substrate scope and significantly more regioselective.[7] Other flavin-dependent tryptophan halogenases include tryptophan 5-halogenase and tryptophan 6-halogenase.[8]

Structure
[edit]
Tryptophan 7-halogenase is a 538-residue, 61-kDa protein.[1][2] In solution, a number of homologues exist as homodimers.[1]
The enzyme has two main binding sites: one for FAD and one for tryptophan. Note that the FAD is supplied by a separate flavin reductase which may be a general enzyme recruited from metabolism or a specific enzyme encoded in the relevant biosynthetic gene cluster.[9] The former site is also the site of reaction for the oxidation of FAD by O2. Connecting these two sites is a tunnel-like structure approximately 10 Å in length. The FAD binding site also binds a single chloride ion.[10] A chlorinating agent is generated at the FAD-binding site, then channeled through this tunnel where it reacts with the substrate tryptophan. Tryptophan is bound by a number of interactions: other aromatic amino acid residues such as tryptophan, phenylalanine, and histidine seemingly "sandwich" the substrate, the N-H group of tryptophan forms hydrogen bonding interactions with the peptide backbone of the enzyme, and the amino acid moiety of tryptophan interacts with nearby tyrosine and glutamate residues.[10] It is thought that the regioselectivity of the enzyme is a consequence of the spatial orientation of the tryptophan, with C7 placed close to the end of the tunnel where the chlorinating agent exits.[11] Multiple halogenations are prevented as the addition of a halogen atom to the substrate greatly increases its steric bulk, causing it to dissociate from the enzyme.[10]
Mechanism
[edit]
Tryptophan 7-halogenases are FADH2-dependent, meaning they require an FADH2 cofactor in order to carry out their reaction. Flavin reductases are responsible for the conversion of FAD to FADH2. For sustained activity in an in vitro setting, tryptophan 7-halogenases thus require either excess FADH2 or the presence of a flavin reductase.[3] Since flavin reductase is itself NAD(P)H-dependent, a recent work studying RebH used a cofactor regeneration system wherein glucose dehydrogenase reduces NAD(P)+ to NAD(P)H, which RebF (RebH's native flavin reductase partner) uses to reduce FAD to FADH2 for subsequent use in RebH’s active site.[12] Mechanistically, the flavin-dependent halogenases are similar to flavin-containing monooxygenases. In the first step, FADH2 is oxidized by molecular oxygen, generating a high-energy flavin hydroperoxide.[9]
At this point, there are two proposed mechanisms to achieve halogenation.[2]
In the first, flavin hydroperoxide attacks the C6-C7 double bond of tryptophan, generating an epoxide. This epoxide is then opened by regioselective nucleophilic attack of halide anion at C7. The resultant halohydrin is dehydrated to form 7-chlorotryptophan.[13] This mechanism is dubious due to the long channel separating the flavin cofactor from the tryptophan substrate.

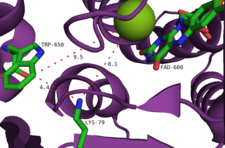
Another proposed mechanism involves the interception of flavin hydroperoxide by a halide anion, generating an equivalent of hypohalous acid. It is unlikely that this species is the final halogenating agent, as experiments have suggested that there is no free hypohalous acid generated by the enzyme.[2] Rather, it is proposed that the 10Å tunnel within the active site directs HOCl toward a lysine residue to form a relatively long lived chloramine intermediate (half-life ~26 hours).[14] Notably, this lysine residue (Lys79) is conserved in all flavin-dependent halogenases.[10] Chloramine, though a weaker halogenating agent, is far more selective.[9]
Studies also indicate that a neighboring glutamate residue is necessary for activity - it is likely that the glutamate residue works in tandem with the lysine residue to generate a more appropriate halogenating agent than a hypohalous acid.[10] This glutamate residue is conserved in all flavin-dependent tryptophan halogenases as well.[10] Proposed roles for this glutamate include a weak interaction with the proton of hypohalous acid to increase the electrophilicity of the chlorine atom or a polarization of the chlorine atom (whether the true chlorine species is hypohalous acid or chloramine) through glutamate's negatively charged carboxylate group.[11]
Spectroscopic and kinetic studies tend to support this mechanism. Flavin redox chemistry was shown to be complete before chlorination; in addition, it can occur without tryptophan present, lending credence to the idea that chloride oxidation and substrate chlorination are decoupled processes.[15] Radiolabeling with 36Cl shows that the chlorine atom goes through three states in this halogenation: free ion, protein-bound, and substrate-bound. This data is in agreement with the proposed mechanism in which lysine forms a covalent adduct with the chlorine species.[14]
In either case, the flavin hydroperoxide transfers one oxygen to the reactants. With elimination of water, FAD is restored.
Biological relevance
[edit]Numerous medicinally relevant compounds depend upon tryptophan 7-halogenase in their biosynthesis. One notable example is the biosynthesis of the antifungal antibiotic pyrrolnitrin in Pseudomonas fluorescens - it was in this context that tryptophan 7-halogenase was first isolated.[7][10] Also, in Lechevalieria aerocolonigenes, the enzyme catalyses the initial step in the biosynthesis of antitumor agent rebeccamycin.[16]

Industrial relevance
[edit]In numerous industries such as agrochemicals and pharmaceuticals, halogenated chemicals abound. Aryl halides are also important intermediates in the syntheses of other compounds due to their versatility in reactions such as nucleophilic aromatic substitution and coupling reactions.[8] Traditionally, in chemical synthesis, arenes are halogenated by electrophilic aromatic substitution, a process that often suffers from poor regioselectivity and makes use of toxic reagents and catalysts under harsh reaction conditions. Tryptophan 7-halogenase circumvents these issues, halogenating its substrate regioselectively and using halide salts and molecular oxygen as reagents at room temperature and fairly neutral pH.[17] Consequently, much research has been dedicated to modifying characteristics of tryptophan 7-halogenase to change properties such as substrate scope, optimal operating temperature, thermal stability, regioselectivity, and more.
Directed evolution is a particularly popular methodology to achieve desirable properties. As of yet, tryptophan 7-halogenase has not been isolated from any thermophilic organism, so augmenting the thermal properties of the enzyme through unnatural means was necessary. As elevated temperatures allow for greater reaction rates, it is desirable to raise an enzyme's optimal operating temperature, but the thermal stability of the enzyme must accordingly be improved to prevent thermal unfolding. Through directed evolution, advances were achieved such as an increase in Topt from 30-35 °C to 40 °C and an increase in Tm from 52.4 °C to 70.0 °C.[18] In combination with directed evolution, substrate walking generated a number of enzymes with greater substrate scope. Enzyme activity was evolved with a known substrate that bears structural similarity to the target substrate, then variants with high activity with respect to the target substrate are starting points for further evolution. Substrates significantly larger than tryptophan were able to be halogenated through this approach. Carvedilol, an indole derivative with a molar mass of over 400 g mol−1 (compared to tryptophan's ~200 g mol−1), was able to be halogenated by an evolved enzyme with high selectivity and yield and low enzyme loading. These results suggest that for larger substrates, perhaps the entirety of the substrate molecule is not enclosed in the active site.[8]
Site-directed mutagenesis has also been employed for expanding substrate scope. Tryptophan substrate interacts with numerous other aromatic amino acid residues near the binding site, poising it for regioselective halogenation. Modification of these residues in turn can allow tryptophan to adopt different spatial orientations that would offer alternative regioselectivities.[10]
Naturally occurring tryptophan 7-halogenase isolated from Lechevalieria aerocolonigenes was shown to be able to halogenate naphthalene derivatives, albeit only at activated positions, revealing that certain strains of tryptophan 7-halogenase are fairly wide in substrate scope.[12]

Following design of a suitable enzyme, a cross-linked enzyme aggregate is used for larger-scale reaction. The enzyme by itself faces stability issues, but immobilization through CLEA circumvents this problem. As tryptophan 7-halogenase requires a number of partner enzymes, a combiCLEA was created, where the halogenase is linked with auxiliary enzymes such as flavin reductase and alcohol dehydrogenase, which are responsible for production of FADH2 and NADH, respectively. The three enzymes were precipitated with ammonium sulfate, then cross-linked with the dialdehyde glutaraldehyde.[8] Low concentrations of glutaraldehyde showed the greatest activity, suggesting the importance of the lysine residue covered in the mechanism discussion, though it is also known that high concentrations of glutaraldehyde in general disrupt enzyme activity, as shown when a separate study was preparing a chloroperoxidase-based CLEA.[19] This combiCLEA allowed for gram-scale halogenation, and the aggregate also offered improved properties over the natural enzyme such as recyclability, long-term storage capability, and ease of purification.[8]
References
[edit]- ^ a b c Dong C, Kotzsch A, Dorward M, van Pée KH, Naismith JH (August 2004). "Crystallization and X-ray diffraction of a halogenating enzyme, tryptophan 7-halogenase, from Pseudomonas fluorescens". Acta Crystallographica Section D. 60 (Pt 8): 1438–40. doi:10.1107/S0907444904012521. PMID 15272170.
- ^ a b c d Dong C, Flecks S, Unversucht S, Haupt C, van Pée KH, Naismith JH (September 2005). "Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination". Science. 309 (5744): 2216–9. doi:10.1126/science.1116510. PMC 3315827. PMID 16195462.
- ^ a b Yeh E, Garneau S, Walsh CT (March 2005). "Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis". Proceedings of the National Academy of Sciences of the United States of America. 102 (11): 3960–5. doi:10.1073/pnas.0500755102. PMC 554827. PMID 15743914.
- ^ Menon, Binuraj R. K.; Richmond, Daniel; Menon, Navya (2020-10-15). "Halogenases for biosynthetic pathway engineering: Toward new routes to naturals and non-naturals". Catalysis Reviews: 1–59. doi:10.1080/01614940.2020.1823788. ISSN 0161-4940.
- ^ van Pée KH (April 2001). "Microbial biosynthesis of halometabolites". Archives of Microbiology. 175 (4): 250–8. doi:10.1007/s002030100263. PMID 11382220. S2CID 44612286.
- ^ Dairi T, Nakano T, Aisaka K, Katsumata R, Hasegawa M (June 1995). "Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline". Bioscience, Biotechnology, and Biochemistry. 59 (6): 1099–106. doi:10.1271/bbb.59.1099. PMID 7612997.
- ^ a b Burd VN, Van Pee KH (October 2003). "Halogenating enzymes in the biosynthesis of antibiotics". Biochemistry. Biokhimiia. 68 (10): 1132–5. doi:10.1023/a:1026362729567. PMID 14616084. S2CID 29194538.
- ^ a b c d e Frese M, Sewald N (January 2015). "Enzymatic halogenation of tryptophan on a gram scale". Angewandte Chemie. 54 (1): 298–301. doi:10.1002/anie.201408561. PMID 25394328.
- ^ a b c Neumann CS, Fujimori DG, Walsh CT (February 2008). "Halogenation strategies in natural product biosynthesis". Chemistry & Biology. 15 (2): 99–109. doi:10.1016/j.chembiol.2008.01.006. PMID 18291314.
- ^ a b c d e f g h Lang A, Polnick S, Nicke T, William P, Patallo EP, Naismith JH, van Pée KH (March 2011). "Changing the regioselectivity of the tryptophan 7-halogenase PrnA by site-directed mutagenesis". Angewandte Chemie. 50 (13): 2951–3. doi:10.1002/anie.201007896. PMID 21404376.
- ^ a b Flecks S, Patallo EP, Zhu X, Ernyei AJ, Seifert G, Schneider A, Dong C, Naismith JH, van Pée KH (2008-01-01). "New insights into the mechanism of enzymatic chlorination of tryptophan". Angewandte Chemie. 47 (49): 9533–6. doi:10.1002/anie.200802466. PMC 3326536. PMID 18979475.
- ^ a b Payne JT, Andorfer MC, Lewis JC (May 2013). "Regioselective arene halogenation using the FAD-dependent halogenase RebH". Angewandte Chemie. 52 (20): 5271–4. doi:10.1002/anie.201300762. PMC 3824154. PMID 23592388.
- ^ Keller S, Wage T, Hohaus K, Hölzer M, Eichhorn E (July 2000). "Purification and Partial Characterization of Tryptophan 7-Halogenase (PrnA) from Pseudomonas fluorescens". Angewandte Chemie. 39 (13): 2300–2302. doi:10.1002/1521-3773(20000703)39:13<2300::aid-anie2300>3.0.co;2-i. PMID 10941070.
- ^ a b Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT (February 2007). "Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases". Biochemistry. 46 (5): 1284–92. doi:10.1021/bi0621213. PMID 17260957.
- ^ Yeh E, Cole LJ, Barr EW, Bollinger JM, Ballou DP, Walsh CT (June 2006). "Flavin redox chemistry precedes substrate chlorination during the reaction of the flavin-dependent halogenase RebH". Biochemistry. 45 (25): 7904–12. doi:10.1021/bi060607d. PMID 16784243.
- ^ Bitto E, Huang Y, Bingman CA, Singh S, Thorson JS, Phillips GN (January 2008). "The structure of flavin-dependent tryptophan 7-halogenase RebH". Proteins. 70 (1): 289–93. doi:10.1002/prot.21627. PMID 17876823.
- ^ Payne JT, Poor CB, Lewis JC (March 2015). "Directed evolution of RebH for site-selective halogenation of large biologically active molecules". Angewandte Chemie. 54 (14): 4226–30. doi:10.1002/anie.201411901. PMC 4506780. PMID 25678465.
- ^ Poor CB, Andorfer MC, Lewis JC (June 2014). "Improving the stability and catalyst lifetime of the halogenase RebH by directed evolution". ChemBioChem. 15 (9): 1286–9. doi:10.1002/cbic.201300780. PMC 4124618. PMID 24849696.
- ^ Perez DI, van Rantwijk F, Sheldon RA (September 2009). "Cross-Linked Enzyme Aggregates of Chloroperoxidase: Synthesis, Optimization and Characterization". Advanced Synthesis & Catalysis. 351 (13): 2133–2139. doi:10.1002/adsc.200900303.
External links
[edit] Media related to Tryptophan 7-halogenase at Wikimedia Commons
Media related to Tryptophan 7-halogenase at Wikimedia Commons- Tryptophan+7-halogenase at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

