Transmembrane protein 251
Transmembrane protein 251, also known as C14orf109 or UPF0694, is a protein that in humans is encoded by the TMEM251 gene.[1] One notable feature of this protein is the presence of proline residues on one of its predicted transmembrane domains.,[2] which is a determinant of the intramitochondrial sorting of inner membrane proteins.[3]
Gene
[edit]The TMEM251 gene is located on human chromosome 14, at 14q32.12, on the plus strand.[4] The gene size is 1,277 base pairs. It contains 3 distinct introns, and transcription produces six different mRNAs that appear to differ by truncation of the 3' end. There are 2 transcript variants that encode for the TMEM251 protein, with the longer one being 169 base pairs in length, and the shorter one being 131 base pairs in length. The first transcript variant encodes a shorter predicted protein, while the second transcript variant encodes a protein with a longer N-terminus. Both consists of two exons that include the entire coding sequence for the TMEM251 protein.[4]
Figure 1: Chromosome 14 overview. TMEM251 is positioned at 14q32.12, marked by a red line.
Promoter
[edit]According to Genomatix's ElDorado program, the promoter region of TMEM251 is predicted to be 680 base pairs in length. The promoter region starts 500 base pairs upstream of the 5’ UTR of TMEM251 mRNA transcript and contains part of this 5’ UTR.[5]
Transcription Factors
[edit]Various transcription factors are predicted to bind within the conserved parts of the promoter (upstream regulatory) region, on both the plus and minus strands. The transcription factors with the highest matrix scores include NKX homeodomain factors, GATA-binding factors, two-handed zinc finger, E2F transcription factor, and T-box transcription factors. No vertebrate TATA binding protein factors, RNA polymerase transcription factor II B, CCAAT binding factors, or CCAAT enhancer binding proteins were found.[6]
Protein
[edit]The TMEM251 protein is 169 amino acids in length. The molecular weight of this protein is 18,747 daltons, with an isoelectric point of 8.38.[1] It is known to be a type IV multi-pass membrane because it spans the membrane twice in alpha-helical configuration, with its N-terminal domains targeted to the lumen.[7] The TMEM251 protein contains a domain of unknown function, part of the domain family DUF4583, spanning from amino acids 35-160. TMEM251 has two isoforms, TMEM251.1 and TMEM251.2.[4]
Protein Composition
[edit]Leucine is the most abundant amino acid by volume (15.37%). TMEM251 has very low abundance of Cysteine, Asparagine, and Aspartic acid. It has one negative charge cluster from amino acid 67–82. No repeats are identified. The same patterns are observed in TMEM251's primate orthologs.[8]
Tissue Expression
[edit]In the human body, microarray-assessed tissue expression patterns show TMEM251 to be highly expressed in ascites, bladder, bone, embryonic tissue, intestine, and skin. In terms of clinical relevance, TMEM251 is expressed in breast carcinoma, dendritic cell line, hepatocellular carcinoma, neuroblastoma, glioblastoma, adult B-acute lymphoblastic leukemia, and blood mononuclear tissues (75-98%). Over-expression of the TMEM251 gene has not been linked as a causal factor in any of these disease states[9]
The conditions under which TMEM251 rises include occupational benzene exposure, acute cold exposure, macular degeneration and dermal fibroblast, and asthma. These microarray-assessed samples have low percentage rank on NCBI Geo (mostly below 50%). The conditions under which TMEM251 falls include infantile-onset Pompe disease, caseous tuberculosis granulomas, and endurance exercise training. These samples have relatively high percentage rank (mostly above 70%).[9]
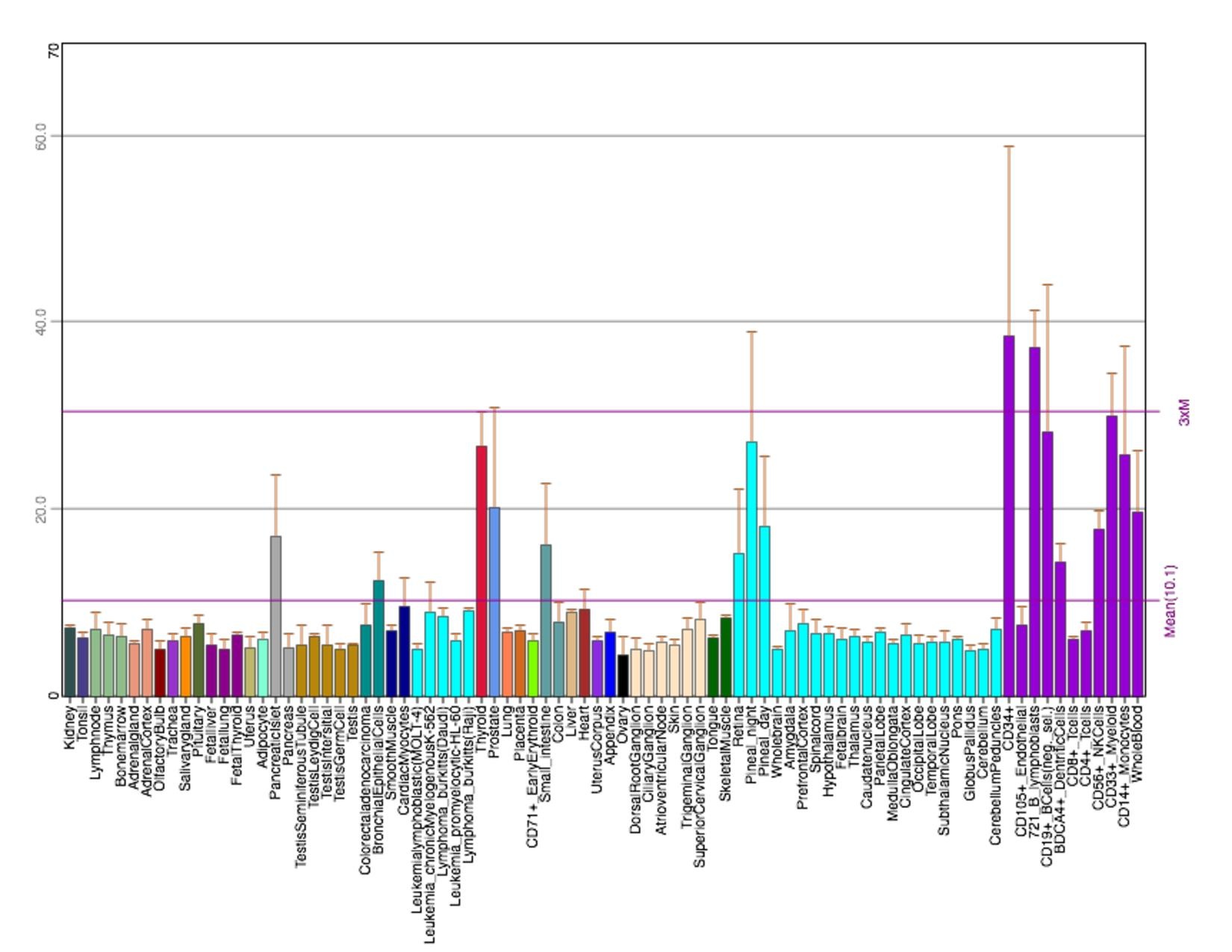
Figure 2: EST Profile data shows the tissue expression of TMEM251 in humans.[10]
Homology and Evolution
[edit]TMEM251 has no paralogs in humans. It does have orthologs within eukaryotes. Conservation has only been found in primates, not in bacteria, plants, or fungus. The following table represents a small selection of orthologs found using searches in BLAST[11] and BLAT,[8] sorted by % identity. This is by no means a comprehensive list, however it does show the vast diversity of species where TMEM251 orthologs are found.
| Genus and Species | Common Name | Date of Divergence | Length | Identity | E-value | Notes |
|---|---|---|---|---|---|---|
| Pan troglodytes | Chimpanzee | 6.3 MYA | 169aa | 99% | 1e-121 | 5’ and 3’ are not truncated |
| Nomascus leucogenys | Gibbon | 20.4 MYA | 169aa | 97% | 1e-119 | 5’ truncated |
| Pteropus alecto | Black flying bat | 94.2 MYA | 175aa | 97% | 1e-119 | 5’ truncated |
| Dasypus novemcinctus | Armadillo | 104.2 MYA | 163aa | 97% | 3e-115 | 5’ truncated |
| Canis lupus familiaris | Dog | 94.2 MYA | 163aa | 97% | 7e-115 | 5’ truncated |
| Odobenus rosmarus divergens | Walrus | 94.2 MYA | 169aa | 96% | 3e-118 | 5’ truncated |
| Ictidomys tridecemlineatus | Ground squirrel | 92.3 MYA | 169aa | 96% | 3e-118 | 5’ truncated |
| Tinamus guttatus | White-throated tinamou | 296 MYA | 131aa | 92% | 1e-85 | 5’ truncated |
| Xenopus (Silurana) tropicalis | Western clawed frog | 371.2 MYA | 130aa | 85% | 2e-81 | 5’ truncated |
| Corvus cornix cornix | Hooded crow | 296 MYA | 171aa | 84% | 4e-91 | 5’ truncated |
| Danio rerio | Zebrafish | 141aa | 141aa | 69% | 9e-63 | 5’ truncated |
The TMEM251 gene first appeared on the planet around 400 million years ago (MYA), since the most distant orthologs are found in fish which diverged from humans around the same time. The size of the gene family, which is a set of similar genes that are formed by duplication of an original gene, is around 120 genes. Gene duplication, resulting in paralogous genes, occurred approximately 371.2 million years ago.[8]
Post-Translational Modifications
[edit]Using various tools at ExPASy, the following are possible post-translational modifications for TMEM251:
- 3 possible Serine phosphorylation sites, no Threonine or Tyrosine phosphorylation sites.[12]
- PKC phosphorylation site on Threonine-26.[13]
- N-terminal acetylation site at the A position of –MLAFSE.[14]
- SUMO interaction site on amino acids 139–143.[15]
- 4 O-beta-N-acetylglucosamine attachment sites
All post-translational modifications are conserved in vertebrates.[2]
Protein Secondary Structure
[edit]Using various tools at ExPASy, TMEM251 secondary structure consists of the following:
- 74.6% Beta-sheet.
- 71.6% alpha helix.
- 8.9% turns
It is predicted to have two transmembrane helices, of 23 amino acids in length each. The average hydrophobicity is predicted to be 0.19.[2]
Figure 3: TMEM251 predicted secondary structure from SOSUI.[16]
Mutation
[edit]TMEM251 has a multitude of mutations in its 5'UTR, coding sequence, and 3'UTR. The majority of the mutations observed are missense mutations.[17]
References
[edit]- ^ a b "TMEM251". GeneCards. LifeMap Sciences. Retrieved 9 May 2015.
- ^ a b c "ExPASy". ExPASy. SIB Bioinformatics Resource Portal. Retrieved 9 May 2015.
- ^ Meier, Stephan; Neupert, Walter; Herrmann, Johannes M. (2005). "Proline residues of transmembrane domains determine the sorting of inner membrane proteins in mitochondria". The Journal of Cell Biology. 170 (6): 881–888. doi:10.1083/jcb.200505126. PMC 2171449. PMID 16157698.
- ^ a b c "TMEM251 transmembrane protein 251 [Homo sapiens (human)]". NCBI Gene. U.S. National Library of Medicine. Retrieved 9 May 2015.
- ^ "Genomes and Annotation: ElDorado". ElDorado. Genomatix. Retrieved 9 May 2015.
- ^ "MatInspector". Genomatix. Genomatix. Retrieved 10 May 2015.
- ^ "Q8N6I4 - TM251_HUMAN". UniProt. UnitProt. Retrieved 9 May 2015.
- ^ a b c "Human BLAT Search". UCSC Genome Browser. UCSC. Retrieved 9 May 2015.
- ^ a b "Gene Expression Omnibus". NCBI Geo. NCBI Geo. Retrieved 9 May 2015.
- ^ "BioGPS". BioGPS. BioGPS. Retrieved 9 May 2015.
- ^ "Basic Local Alignment Search Tool". NCBI Blast. NCBI. Retrieved 9 May 2015.
- ^ "NetPhos 2.0 Server". NetPhos 2.0 Server. Center of Biological Sequence Analysis. Retrieved 9 May 2015.
- ^ "NetPhosK 1.0 Server". NetPhosK 1.0 Server. Center of Biological Sequence Analysis. Retrieved 9 May 2015.
- ^ "NetAcet 1.0 Server". NetAcet 1.0 Server. Center of Biological Sequence Analysis. Retrieved 9 May 2015.
- ^ "GPS-SUMO: Prediction of SUMOylation Sites". GPS-SUMO: Prediction of SUMOylation Sites. SUMOsp. Retrieved 9 May 2015.
- ^ "SOSUI". SOSUI. Harrier Nagahama. Retrieved 10 May 2015.
- ^ "NCBI dbSNP". NCBI. National Center for Biotechnology Information. Retrieved 10 May 2015.

