RNA recognition motif
| RNA recognition motif. (a.k.a. RRM, RBD, or RNP domain) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | RRM_1 | ||||||||||
| Pfam | PF00076 | ||||||||||
| Pfam clan | CL0221 | ||||||||||
| ECOD | 304.9.1 | ||||||||||
| InterPro | IPR000504 | ||||||||||
| PROSITE | PDOC00030 | ||||||||||
| SCOP2 | 1sxl / SCOPe / SUPFAM | ||||||||||
| |||||||||||
RNA recognition motif, RNP-1 is a putative RNA-binding domain of about 90 amino acids that are known to bind single-stranded RNAs. It was found in many eukaryotic proteins.[1][2][3]
The largest group of single strand RNA-binding protein is the eukaryotic RNA recognition motif (RRM) family that contains an eight amino acid RNP-1 consensus sequence.[4][5]
RRM proteins have a variety of RNA binding preferences and functions, and include heterogeneous nuclear ribonucleoproteins (hnRNPs), proteins implicated in regulation of alternative splicing (SR, U2AF2, Sxl), protein components of small nuclear ribonucleoproteins (U1 and U2 snRNPs), and proteins that regulate RNA stability and translation (PABP, La, Hu).[2][3][5] The RRM in heterodimeric splicing factor U2 snRNP auxiliary factor appears to have two RRM-like domains with specialised features for protein recognition.[6] The motif also appears in a few single stranded DNA binding proteins.
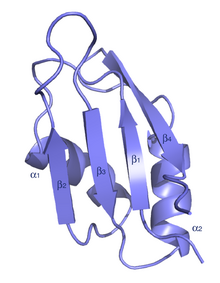
The typical RRM consists of four anti-parallel beta-strands and two alpha-helices arranged in a beta-alpha-beta-beta-alpha-beta fold with side chains that stack with RNA bases. A third helix is present during RNA binding in some cases.[7] The RRM is reviewed in a number of publications.[8][9][10]
Human proteins containing this domain
[edit]A2BP1; ACF; BOLL; BRUNOL4; BRUNOL5; BRUNOL6; CCBL2; CGI-96; CIRBP; CNOT4; CPEB2; CPEB3; CPEB4; CPSF7; CSTF2; CSTF2T; CUGBP1; CUGBP2; D10S102; DAZ1; DAZ2; DAZ3; DAZ4; DAZAP1; DAZL; DNAJC17; DND1; EIF3S4; EIF3S9; EIF4B; EIF4H; ELAVL1; ELAVL2; ELAVL3; ELAVL4; ENOX1; ENOX2; EWSR1; FUS; FUSIP1; G3BP; G3BP1; G3BP2; GRSF1; HNRNPL; HNRPA0; HNRPA1; HNRPA2B1; HNRPA3; HNRPAB; HNRPC; HNRPCL1; HNRPD; HNRPDL; HNRPF; HNRPH1; HNRPH2; HNRPH3; HNRPL; HNRPLL; HNRPM; HNRPR; HRNBP1; HSU53209; HTATSF1; IGF2BP1; IGF2BP2; IGF2BP3; LARP7; MKI67IP; MSI1; MSI2; MSSP-2; MTHFSD; MYEF2; NCBP2; NCL; NOL8; NONO; P14; PABPC1; PABPC1L; PABPC3; PABPC4; PABPC5; PABPN1; POLDIP3; PPARGC1; PPARGC1A; PPARGC1B; PPIE; PPIL4; PPRC1; PSPC1; PTBP1; PTBP2; PUF60; RALY; RALYL; RAVER1; RAVER2; RBM10; RBM11; RBM12; RBM12B; RBM14; RBM15; RBM15B; RBM16; RBM17; RBM18; RBM19; RBM22; RBM23; RBM24; RBM25; RBM26; RBM27; RBM28; RBM3; RBM32B; RBM33; RBM34; RBM35A; RBM35B; RBM38; RBM39; RBM4; RBM41; RBM42; RBM44; RBM45; RBM46; RBM47; RBM4B; RBM5; RBM7; RBM8A; RBM9; RBMS1; RBMS2; RBMS3; RBMX; RBMX2; RBMXL2; RBMY1A1; RBMY1B; RBMY1E; RBMY1F; RBMY2FP; RBPMS; RBPMS2; RDBP; RNPC3; RNPC4; RNPS1; ROD1; SAFB; SAFB2; SART3; SETD1A; SF3B6; SF3B4; SFPQ; SFRS1; SFRS10; SFRS11; SFRS12; SFRS15; SRSF2; SFRS2B; SFRS3; SFRS4; SFRS5; SFRS6; SFRS7; SFRS9; SLIRP; SLTM; SNRP70; SNRPA; SNRPB2; SPEN; SR140; SRRP35; SSB; SYNCRIP; TAF15; TARDBP; THOC4; TIA1; TIAL1; TNRC4; TNRC6C; TRA2A; TRSPAP1; TUT1; U1SNRNPBP; U2AF1; U2AF2; UHMK1; ZCRB1; ZNF638; ZRSR1; ZRSR2;
References
[edit]- ^ Swanson MS, Dreyfuss G, Pinol-Roma S (1988). "Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation". Trends Biochem. Sci. 13 (3): 86–91. doi:10.1016/0968-0004(88)90046-1. PMID 3072706.
- ^ a b Keene JD, Chambers JC, Kenan D, Martin BJ (1988). "Genomic structure and amino acid sequence domains of the human La autoantigen". J. Biol. Chem. 263 (34): 18043–51. doi:10.1016/S0021-9258(19)81321-2. PMID 3192525.
- ^ a b Davis RW, Sachs AB, Kornberg RD (1987). "A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability". Mol. Cell. Biol. 7 (9): 3268–76. doi:10.1128/mcb.7.9.3268. PMC 367964. PMID 3313012.
- ^ Bandziulis RJ, Swanson MS, Dreyfuss G (1989). "RNA-binding proteins as developmental regulators". Genes Dev. 3 (4): 431–437. doi:10.1101/gad.3.4.431. PMID 2470643.
- ^ a b Keene JD, Query CC, Bentley RC (1989). "A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein". Cell. 57 (1): 89–101. doi:10.1016/0092-8674(89)90175-X. PMID 2467746. S2CID 22127152.
- ^ Green MR, Kielkopf CL, Lucke S (2004). "U2AF homology motifs: protein recognition in the RRM world". Genes Dev. 18 (13): 1513–1526. doi:10.1101/gad.1206204. PMC 2043112. PMID 15231733.
- ^ Kumar S, Birney E, Krainer AR (1993). "Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors". Nucleic Acids Res. 21 (25): 5803–5816. doi:10.1093/nar/21.25.5803. PMC 310458. PMID 8290338.
- ^ Keene JD, Kenan DJ, Query CC (1991). "RNA recognition: towards identifying determinants of specificity". Trends Biochem. Sci. 16 (6): 214–20. doi:10.1016/0968-0004(91)90088-d. PMID 1716386.
- ^ Allain FH, Dominguez C, Maris C (2005). "The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression". FEBS J. 272 (9): 2118–31. doi:10.1111/j.1742-4658.2005.04653.x. PMID 15853797. S2CID 46680279.
- ^ Teplova M, Yuan YR, Patel DJ, Malinina L, Teplov A, Phan AT, Ilin S (2006). "Structural basis for recognition and sequestration of UUU(OH) 3' temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen". Mol. Cell. 21 (1): 75–85. doi:10.1016/j.molcel.2005.10.027. PMC 4689297. PMID 16387655.
External links
[edit]- Eukaryotic Linear Motif resource motif class LIG_ULM_U2AF65_1
