NT5C3
| NT5C3A | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | NT5C3A, NT5C3, P5'N-1, P5N-1, PN-I, POMP, PSN1, UMPH, UMPH1, cN-III, hUMP1, p36, 5'-nucleotidase, cytosolic IIIA | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 606224; MGI: 1927186; HomoloGene: 9534; GeneCards: NT5C3A; OMA:NT5C3A - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| EC number | 3.1.3.91 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Cytosolic 5'-nucleotidase 3 (NTC53), also known as cytosolic 5'-nucleotidase 3A, pyrimidine 5’-nucleotidase (PN-I or P5'NI), and p56, is an enzyme that in humans is encoded by the NT5C3, or NT5C3A, gene on chromosome 7.[5][6][7][8]
This gene encodes a member of the 5'-nucleotidase family of enzymes that catalyze the dephosphorylation of nucleoside 5'-monophosphates. The encoded protein is the type 1 isozyme of pyrimidine 5' nucleotidase and catalyzes the dephosphorylation of pyrimidine 5' monophosphates. Mutations in this gene are a cause of hemolytic anemia due to uridine 5-prime monophosphate hydrolase deficiency. Alternatively spliced transcript variants encoding multiple isoforms have been observed for this gene, and pseudogenes of this gene are located on the long arm of chromosomes 3 and 4. [provided by RefSeq, Mar 2012][7]
Structure
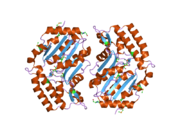
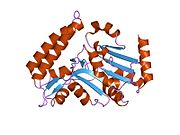
[edit]The NT5C3 gene consists of 10 exons and can be alternatively spliced at exon 2.[9] Four possible isoforms have been identified, encoding proteins with lengths of 336 residues, 297 residues, 286 residues, and 285 residues.[9][10] The 286-residue long isozyme is a monomeric protein containing 5 cysteine residues and no disulfide bridges or phosphate content.[8][9] It has a predicted mass of 32.7 kDa and a predicted globular tertiary structure consisting of approximately 30% α-helices and 26% extended strands.[9] This enzyme structurally resembles members of the haloacid dehalogenase (HAD) superfamily in regards to the shared α/β-Rossmann-like domain and a smaller 4-helix bundle domain. Three motifs in the α/β-Rossmann-like domain form the catalytic phosphate-binding site. Motif I is responsible for the 5′-nucleotidase activity: the first Asp makes a nucleophilic attack on the phosphate of the nucleoside monophosphate substrate, while the second Asp donates a proton to the leaving nucleoside. The active site is located in a cleft between the α/β-Rossmann-like domain and 4-helix bundle domain.[11]
Function
[edit]NT5C3 is a member of the 5'-nucleotidase family and one of the five cytosolic members identified in humans.[10] NTC53 catalyzes the dephosphorylation of the pyrimidine 5′ monophosphates UMP and CMP to the corresponding nucleosides.[8][9] This function contributes to RNA degradation during the erythrocyte maturation process.[6][8][10] As a result, NT5C3 regulates both the endogenous nucleoside and nucleotide pool balance, as well as that of pyrimidine analogs such as gemcitabine and AraC.[10]
NT5C3 was first discovered in red blood cells, but its expression has been observed in multiple tumors (lung, ovary, colon, bladder), fetal tissues (lung, heart, spleen, liver), adult testis, and the brain.[6][9] In particular, the 297-residue isoform of this enzyme is highly expressed in lymphoblastoid cells.[10]
Clinical Significance
[edit]The loss of NT5C3 in pyrimidine 5' nucleotidase deficiency, an autosomal recessive condition, leads to the accumulation of high concentrations of pyrimidine nucleotides within erythrocytes.[6][8][9] This deficiency is characterized by moderate hemolytic anemia, jaundice, splenomegaly, and marked basophilic stippling, and has been associated with learning difficulties.[6][9] Two homozygous mutations identified in this gene produced large deletions that could cripple the enzyme’s structure and function, and are thus causally linked to pyrimidine 5' nucleotidase deficiency and hemolytic anemia. Heterozygous mutations in pyrimidine 5' nucleotidase deficiency may contribute to the large variability in thalassemia phenotypes.[9] Pyrimidine 5' nucleotidase deficiency is also linked to the conversion of hemoglobin E disease into an unstable hemoglobinopathy-like disease.[6][9] NT5C3 is identical to p36, a previously identified alpha-interferon-induced protein involved in forming lupus inclusions.[6][8] Since NT5C3 can metabolize AraC, a nucleoside analog used in chemotherapy for acute myeloid leukemia patients, genotyping one of its polymorphisms may aid detection of patients who will respond favorably to this therapy.[12]
Interactions
[edit]NTC53 is known to interact with pyrimidine nucleoside monophosphates, specifically UMP and CMP, as well as the anineoplastic agents 5’-AZTMP and 5’-Ara-CMP.[8]
References
[edit]- ^ a b c GRCh38: Ensembl release 89: ENSG00000122643 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000029780 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Zhang QH, Ye M, Wu XY, Ren SX, Zhao M, Zhao CJ, Fu G, Shen Y, Fan HY, Lu G, Zhong M, Xu XR, Han ZG, Zhang JW, Tao J, Huang QH, Zhou J, Hu GX, Gu J, Chen SJ, Chen Z (Nov 2000). "Cloning and Functional Analysis of cDNAs with Open Reading Frames for 300 Previously Undefined Genes Expressed in CD34+ Hematopoietic Stem/Progenitor Cells". Genome Res. 10 (10): 1546–60. doi:10.1101/gr.140200. PMC 310934. PMID 11042152.
- ^ a b c d e f g Amici A, Emanuelli M, Raffaelli N, Ruggieri S, Saccucci F, Magni G (Sep 2000). "Human erythrocyte pyrimidine 5-nucleotidase, PN-I, is identical to p36, a protein associated to lupus inclusion formation in response to alpha-interferon". Blood. 96 (4): 1596–8. doi:10.1182/blood.V96.4.1596. PMID 10942414.
- ^ a b "Entrez Gene: NT5C3 5'-nucleotidase, cytosolic III".
- ^ a b c d e f g Amici A, Magni G (15 January 2002). "Human erythrocyte pyrimidine 5'-nucleotidase, PN-I". Archives of Biochemistry and Biophysics. 397 (2): 184–90. doi:10.1006/abbi.2001.2676. PMID 11795870.
- ^ a b c d e f g h i j Marinaki AM, Escuredo E, Duley JA, Simmonds HA, Amici A, Naponelli V, Magni G, Seip M, Ben-Bassat I, Harley EH, Thein SL, Rees DC (1 June 2001). "Genetic basis of hemolytic anemia caused by pyrimidine 5' nucleotidase deficiency". Blood. 97 (11): 3327–32. doi:10.1182/blood.v97.11.3327. PMID 11369620.
- ^ a b c d e Aksoy P, Zhu MJ, Kalari KR, Moon I, Pelleymounter LL, Eckloff BW, Wieben ED, Yee VC, Weinshilboum RM, Wang L (August 2009). "Cytosolic 5'-nucleotidase III (NT5C3): gene sequence variation and functional genomics". Pharmacogenetics and Genomics. 19 (8): 567–76. doi:10.1097/fpc.0b013e32832c14b8. PMC 2763634. PMID 19623099.
- ^ Walldén K, Stenmark P, Nyman T, Flodin S, Gräslund S, Loppnau P, Bianchi V, Nordlund P (15 June 2007). "Crystal structure of human cytosolic 5'-nucleotidase II: insights into allosteric regulation and substrate recognition". The Journal of Biological Chemistry. 282 (24): 17828–36. doi:10.1074/jbc.m700917200. PMID 17405878.
- ^ Cheong HS, Koh Y, Ahn KS, Lee C, Shin HD, Yoon SS (September 2014). "NT5C3 polymorphisms and outcome of first induction chemotherapy in acute myeloid leukemia". Pharmacogenetics and Genomics. 24 (9): 436–41. doi:10.1097/fpc.0000000000000072. PMID 25000516. S2CID 21233346.
Further reading
[edit]- Amici A, Emanuelli M, Ferretti E, et al. (1995). "Homogeneous pyrimidine nucleotidase from human erythrocytes: enzymic and molecular properties". Biochem. J. 304 (Pt 3): 987–92. doi:10.1042/bj3040987. PMC 1137429. PMID 7818506.
- Rich SA, Bose M, Tempst P, Rudofsky UH (1996). "Purification, microsequencing, and immunolocalization of p36, a new interferon-alpha-induced protein that is associated with human lupus inclusions". J. Biol. Chem. 271 (2): 1118–26. doi:10.1074/jbc.271.2.1118. PMID 8557639.
- Hillier LD, Lennon G, Becker M, et al. (1997). "Generation and analysis of 280,000 human expressed sequence tags". Genome Res. 6 (9): 807–28. doi:10.1101/gr.6.9.807. PMID 8889549.
- Amici A, Emanuelli M, Magni G, et al. (1998). "Pyrimidine nucleotidases from human erythrocyte possess phosphotransferase activities specific for pyrimidine nucleotides". FEBS Lett. 419 (2–3): 263–7. doi:10.1016/S0014-5793(97)01464-6. PMID 9428647. S2CID 9588121.
- Hartley JL, Temple GF, Brasch MA (2001). "DNA Cloning Using In Vitro Site-Specific Recombination". Genome Res. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Wiemann S, Weil B, Wellenreuther R, et al. (2001). "Toward a Catalog of Human Genes and Proteins: Sequencing and Analysis of 500 Novel Complete Protein Coding Human cDNAs". Genome Res. 11 (3): 422–35. doi:10.1101/gr.GR1547R. PMC 311072. PMID 11230166.
- Marinaki AM, Escuredo E, Duley JA, et al. (2001). "Genetic basis of hemolytic anemia caused by pyrimidine 5' nucleotidase deficiency". Blood. 97 (11): 3327–32. doi:10.1182/blood.V97.11.3327. PMID 11369620.
- Amici A, Magni G (2002). "Human erythrocyte pyrimidine 5'-nucleotidase, PN-I". Arch. Biochem. Biophys. 397 (2): 184–90. doi:10.1006/abbi.2001.2676. PMID 11795870.
- Amici A, Emanuelli M, Ruggieri S, et al. (2003). "Kinetic Evidence for Covalent Phosphoryl-Enzyme Intermediate in Phosphotransferase Activity of Human Red Cell Pyrimidine Nucleotidases". Enzyme Kinetics and Mechanism - Part F: Detection and Characterization of Enzyme Reaction Intermediates. Methods in Enzymology. Vol. 354. pp. 149–59. doi:10.1016/S0076-6879(02)54011-8. ISBN 978-0-12-182257-6. PMID 12418222.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. Bibcode:2002PNAS...9916899M. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Balta G, Gumruk F, Akarsu N, et al. (2003). "Molecular characterization of Turkish patients with pyrimidine 5' nucleotidase-I deficiency". Blood. 102 (5): 1900–3. doi:10.1182/blood-2003-02-0628. PMID 12714505.
- Bianchi P, Fermo E, Alfinito F, et al. (2003). "Molecular characterization of six unrelated Italian patients affected by pyrimidine 5'-nucleotidase deficiency". Br. J. Haematol. 122 (5): 847–51. doi:10.1046/j.1365-2141.2003.04532.x. PMID 12930399. S2CID 8670252.
- Kanno H, Takizawa T, Miwa S, Fujii H (2004). "Molecular basis of Japanese variants of pyrimidine 5'-nucleotidase deficiency". Br. J. Haematol. 126 (2): 265–71. doi:10.1111/j.1365-2141.2004.05029.x. PMID 15238149. S2CID 27921282.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The Status, Quality, and Expansion of the NIH Full-Length cDNA Project: The Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Wiemann S, Arlt D, Huber W, et al. (2004). "From ORFeome to Biology: A Functional Genomics Pipeline". Genome Res. 14 (10B): 2136–44. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Chiarelli LR, Bianchi P, Fermo E, et al. (2005). "Functional analysis of pyrimidine 5'-nucleotidase mutants causing nonspherocytic hemolytic anemia". Blood. 105 (8): 3340–5. doi:10.1182/blood-2004-10-3895. PMID 15604219.
- Mehrle A, Rosenfelder H, Schupp I, et al. (2006). "The LIFEdb database in 2006". Nucleic Acids Res. 34 (Database issue): D415–8. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.