Mobile genetic elements

Mobile genetic elements (MGEs), sometimes called selfish genetic elements,[1] are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome are thought to be MGEs.[2] MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. These mechanisms can also rearrange genes in the host genome generating variation. These mechanism can increase fitness by gaining new or additional functions. An example of MGEs in evolutionary context are that virulence factors and antibiotic resistance genes of MGEs can be transported to share genetic code with neighboring bacteria. However, MGEs can also decrease fitness by introducing disease-causing alleles or mutations.[3] The set of MGEs in an organism is called a mobilome, which is composed of a large number of plasmids, transposons and viruses.[4]
Types
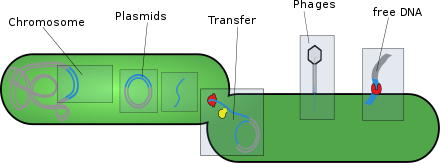
[edit]- Plasmids: These are generally circular extrachromosomal DNA molecules that replicate and are transmitted independently from chromosomal DNA. These molecules are present in prokaryotes (bacteria and archaea) and sometimes in eukaryotic organisms such as yeast. Fitness of a plasmid is determined by its mobility. The first factor of plasmid fitness is its ability to replicate DNA. The second fitness factor is a plasmid's ability to horizontally transfer. Plasmids during their cycle carry genes from one organism to another through a process called conjugation. Plasmids usually contain a set of mobility genes that are necessary for conjugation. Some plasmids employ membrane associated mating pair formation (MPF). A plasmid containing its own MPF genes is considered to be self transmissible or conjugative.[5] Plasmids can be further divided into mobilizable and non-mobilizable classes. Plasmids that use other genetic element MFPs in the cell are mobilizable. Plasmids that are not mobilizable but spread by transduction or transformation are termed non-mobilizable.[5] Plasmids can often inject genes that make bacteria resistant to antibiotics.[6][5]
- Cloning vectors: These are types of hybrid plasmids with bacteriophages, used to transfer and replicate DNA . Fragments of DNA can be inserted by recombinant DNA techniques. A viable vector must be able to replicate together with the DNA fragments it carries. These vectors can contain desired genes for insertion into an organism's genome. Examples are cosmids and phagemids.[7]

- Transposons: These are DNA sequences that can move and replicate in different parts of a cell's genome. Also called "jumping genes", they can be transferred horizontally between organisms that live in symbiosis. Transposons are present in all living things and in giant viruses.[8]
- DNA transposons: These are transposons that move directly from one position to another in the genome using a transposase to cut and stick at another locus.[9] These genetic elements are cleaved at four single stranded sites in DNA by transposase. In order to achieve max stability of the intermediate transposon, one single strand cleavage at the target DNA occurs. Simultaneously the donor strand is ligated to the target strand after cleavage leaving a single strand overhang on either end of the target sequence. These sites usually contain a 5 to 9 base pair overhang that can create a cohesive end.[10] Transposase then holds the sequence in a crossed formation and ligates the donor strand to the target strand. The structure formed by the duplex of DNA and transposase in replicative transposons is known as the Shapiro Intermediate.[11] The 5 to 9 base pair overhang is left on either side of the target sequence allowing it to join to its target sequence in either orientation. The sequence of these overhangs can determine joining orientation.[10] Before site specific recombination can occur, the oligonucleotide ends must be filled. The ligation of these ends generates a replication fork at each end of the transposable element. The single strand displacement causes synthesis from the un-ligated 3' hydroxyl group to form long single stranded sections adjacent to the 5' end. Therefore, the opposite strand is sequenced discontinuously as both replication forks approach the center of the transposable element. This results in two recombinant duplexes containing the semi conserved transposable element flanked by the previous 5 to 9 base pair overhang. Site specific reciprocal recombination takes place between the two transposable elements facilitated by proteins. This reciprocal replication overlaps in time and occurs between duplicated segments of the replication element before replication is completed.[10] The target molecule as a result contains the inserted element flanked by the 5 to 9 base pair sequences. Transposition of these elements duplicates the transposition element leaving a transposition element in its original location and a new transposon at the reciprocal replication site. In doing so, organisms total base pairs in their genomes are increased. Transposition occurrences increase over time and as organisms age.

- Retrotransposons: These are transposons that move in the genome, being transcribed into RNA and later into DNA by reverse transcriptase. Many retrotransposons also exhibit replicative transposition. Retrotransposons are present exclusively in eukaryotes.[12] Retrotransposons consist of two major types, long terminal repeats (LTRs) and Non-LTR transposons. Non-LTR transposons can be further classified into Long interspersed nuclear element (LINEs) and Short interspersed nuclear element (SINEs).[13] These retrotransposons are regulated by a family of short non-coding RNAs termed as PIWI [P-element induced wimpy testis]-interacting RNAs (piRNAs).[14] piRNA is a recently discovered class of ncRNAs, which are in the length range of ~24-32 nucleotides. Initially, piRNAs were described as repeat-associated siRNAs (rasiRNAs) because of their origin from the repetitive elements such as transposable sequences of the genome. However, later it was identified that they acted via PIWI-protein. In addition to having a role in the suppression of genomic transposons, various roles of piRNAs have been recently reported like regulation of 3’ UTR of protein-coding genes via RNAi, transgenerational epigenetic inheritance to convey a memory of past transposon activity, and RNA-induced epigenetic silencing.[14]
- Integrons: These are gene cassettes that usually carry antibiotic resistance genes to bacterial plasmids and transposons.[15]
- Introns: Group I and II introns are nucleotide sequences with catalytic activity that are part of host transcripts and act as ribozymes that can invade genes that encode tRNA, rRNA, and proteins. They are present in all cellular organisms and viruses.[16]
- Introners: Sequences similar to transposons that can jump in the genome leaving new introns where they were, they have been pointed as a possible mechanism of intron gain in the evolution of eukaryotes where they are present in at least 5% of all species, specially in the aquatic taxa due possibly to horizontal gene transfer that occurs more frequently in these animals.[17][18] They were first described in 2009 in the unicellular green algae micromonas.[19]
- Viral agents: These are mostly infective acellular agents that replicate in cellular hosts. During their infective cycle they can carry genes from one host to another. They can also carry genes from one organism to another in case that viral agent infects more than two different species. Traditionally they are considered separate entities, but the truth is that many researchers who study their characteristics and evolution refer to them as mobile genetic elements. This is based on the fact that viral agents are simple particles or molecules that replicate and are transferred between various hosts like the remaining non-viral mobile genetic elements. According to this point of view, viruses and other viral agents should not be considered living beings and should be better conceived as mobile genetic elements. Viral agents are evolutionarily connected with various mobile genetic elements.[20] These viral agents are thought to have arisen from secreted or ejected plasmids of other organisms. Transposons also provide insight into how these elements may have originally started. This theory is known as the vagrancy hypothesis proposed by Barbara McClintock in 1950.[21][1][22][4][23]
- Viruses: These are viral agents composed of a molecule of genetic material (DNA or RNA) and with the ability to form complex particles called virions to be able to move easily between their hosts. Viruses are present in all living things. Viral particles are manufactured by the host's replicative machinery for horizontal transfer.[20][21][24]
- Satellite nucleic acids: These are DNA or RNA molecules, which are encapsulated as a stowaway in the virions of certain helper viruses and which depend on these to be able to replicate. Although they are sometimes considered genetic elements of their helper viruses, they are not always found within their helper viruses.[20][21][25]
- Viroids: These are viral agents that consist of small circular RNA molecules that infect and replicate in plants. These mobile genetic elements do not have a protective protein coating. Specifically, these mobile genetic elements are found in angiosperms.[20][21][26]
- Endogenous viral element: These are viral nucleic acids integrated into the genome of a cell. They can move and replicate multiple times in the host cell without causing disease or mutation. They are considered autonomous forms of transposons. Examples are proviruses and endogenous retroviruses.[27]
Research examples
[edit]CRISPR-Cas systems in bacteria and archaea are adaptive immune systems to protect against deadly consequences from MGEs. Using comparative genomic and phylogenetic analysis, researchers found that CRISPR-Cas variants are associated with distinct types of MGEs such as transposable elements. In CRISPR-associated transposons, CRISPR-Cas controls transposable elements for their propagation.[28]
MGEs such as plasmids by a horizontal transmission are generally beneficial to an organism. The ability of transferring plasmids (sharing) is important in an evolutionary perspective. Tazzyman and Bonhoeffer found that fixation (receiving) of the transferred plasmids in a new organism is just as important as the ability to transfer them.[29] Beneficial rare and transferable plasmids have a higher fixation probability, whereas deleterious transferable genetic elements have a lower fixation probability because they are lethal to the host organisms.
One type of MGEs, namely the Integrative Conjugative Elements (ICEs) are central to horizontal gene transfer shaping the genomes of prokaryotes enabling rapid acquisition of novel adaptive traits.[30][31]
As a representative example of ICEs, the ICEBs1 is well-characterized for its role in the global DNA damage SOS response of Bacillus subtilis[32] and also its potential link to the radiation and desiccation resistance of Bacillus pumilus SAFR-032 spores,[33] isolated from spacecraft cleanroom facilities.[34][35][36]
Transposition by transposable elements is mutagenic. Thus, organisms have evolved to repress the transposition events, and failure to repress the events causes cancers in somatic cells. Cecco et al. found that during early age transcription of retrotransposable elements are minimal in mice, but in advanced age the transcription level increases.[37] This age-dependent expression level of transposable elements is reduced by calorie restriction diet. Replication of transposable elements often results in repeated sequences being added into the genome. These sequences are often non coding but can interfere with coding sequences of DNA. Though mutagenetic by nature, transposons increase the genome of an organism that they transpose into. More research should be conducted into how these elements may serve as a rapid adaptation tool employed by organisms to generate variability. Many transposition elements are dormant or require activation. should also be noted that current values for coding sequences of DNA would be higher if transposition elements that code for their own transposition machinery were considered as coding sequences.
Some others researched examples include Mavericks,[38][39][40] Starships[41][40] and Space invaders (or SPINs)[42][43]
Diseases
[edit]The consequence of mobile genetic elements can alter the transcriptional patterns, which frequently leads to genetic disorders such as immune disorders, breast cancer, multiple sclerosis, and amyotrophic lateral sclerosis. In humans, stress can lead to transactional activation of MGEs such as endogenous retroviruses, and this activation has been linked to neurodegeneration.[44]
Other notes
[edit]The total of all mobile genetic elements in a genome may be referred to as the mobilome.
Barbara McClintock was awarded the 1983 Nobel Prize in Physiology or Medicine "for her discovery of mobile genetic elements" (transposable elements).[45]
Mobile genetic elements play a critical role in the spread of virulence factors, such as exotoxins and exoenzymes, among bacteria. Strategies to combat certain bacterial infections by targeting these specific virulence factors and mobile genetic elements have been proposed.[46]
See also
[edit]- ACLAME (The CLAssification of Mobile genetic Elements) database
- De novo gene birth
- Exon shuffling
- Gene fusion
- Gene duplication
- Horizontal gene transfer
- Virulence factors
- Miniature Inverted-repeat Transposable Elements (MITEs)
References
[edit]- ^ a b Moreira D, López-García P (April 2009). "Ten reasons to exclude viruses from the tree of life". Nature Reviews. Microbiology. 7 (4): 306–311. doi:10.1038/nrmicro2108. PMID 19270719. S2CID 3907750.
- ^ Mu X, Ahmad S, Hur S (2016). Endogenous Retroelements and the Host Innate Immune Sensors. Advances in Immunology. Vol. 132. pp. 47–69. doi:10.1016/bs.ai.2016.07.001. ISBN 9780128047972. PMC 5135014. PMID 27769507.
- ^ Singh PK, Bourque G, Craig NL, Dubnau JT, Feschotte C, Flasch DA, et al. (2014-11-18). "Mobile genetic elements and genome evolution 2014". Mobile DNA. 5: 26. doi:10.1186/1759-8753-5-26. PMC 4363357. PMID 30117500.
- ^ a b Koonin EV, Wolf YI (December 2008). "Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world". Nucleic Acids Research. 36 (21): 6688–6719. doi:10.1093/nar/gkn668. PMC 2588523. PMID 18948295.
- ^ a b c Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F (September 2010). "Mobility of plasmids". Microbiology and Molecular Biology Reviews. 74 (3): 434–452. doi:10.1128/MMBR.00020-10. PMC 2937521. PMID 20805406.
- ^ Summers D (1996). "Chapter 1 – The Function and Organization of Plasmids". The Biology of Plasmids (First ed.). Wiley-Blackwell. pp. 21–22. ISBN 978-0632034369.
- ^ Glick BR, Pasternak JJ (2005). Molecular Biotechnology Principles and Applications of Recombinant DNA (3rd ed.). ASM Press. ISBN 9781555816124.
- ^ Makałowski W, Gotea V, Pande A, Makałowska I (2019). "Transposable Elements: Classification, Identification, and Their Use as a Tool for Comparative Genomics". In Anisimova M (ed.). Evolutionary Genomics. Methods in Molecular Biology. Vol. 1910. New York, NY: Humana. pp. 185–186. doi:10.1007/978-1-4939-9074-0_6. ISBN 978-1-4939-9074-0. PMID 31278665. S2CID 195814061.
- ^ Muñoz-López M, García-Pérez JL (April 2010). "DNA transposons: nature and applications in genomics". Current Genomics. 11 (2): 115–128. doi:10.2174/138920210790886871. PMC 2874221. PMID 20885819.
- ^ a b c Shapiro JA (April 1979). "Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements". Proceedings of the National Academy of Sciences of the United States of America. 76 (4): 1933–1937. Bibcode:1979PNAS...76.1933S. doi:10.1073/pnas.76.4.1933. PMC 383507. PMID 287033.
- ^ Bushman F (2002). Lateral DNA transfer : mechanisms and consequences. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. ISBN 0-87969-603-6. OCLC 47283049.
- ^ Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV (April 2015). "The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes". Microbiology Spectrum. 3 (2): 1165–1208. doi:10.1128/microbiolspec.mdna3-0061-2014. ISBN 9781555819200. PMC 4498412. PMID 26104698.
- ^ Makałowski W, Gotea V, Pande A, Makałowska I (2019). "Transposable Elements: Classification, Identification, and Their Use as a Tool for Comparative Genomics". In Anisimova M (ed.). Evolutionary Genomics. Methods in Molecular Biology. Vol. 1910. New York, NY: Humana. pp. 177–207. doi:10.1007/978-1-4939-9074-0_6. ISBN 978-1-4939-9074-0. PMID 31278665. S2CID 195814061.
- ^ a b Monga I, Banerjee I (November 2019). "Computational Identification of piRNAs Using Features Based on RNA Sequence, Structure, Thermodynamic and Physicochemical Properties". Current Genomics. 20 (7): 508–518. doi:10.2174/1389202920666191129112705. PMC 7327968. PMID 32655289.
- ^ Kovalevskaya NP (2002). "Mobile Gene Cassettes and Integrons". Molecular Biology. 36 (2): 196–201. doi:10.1023/A:1015361704475. S2CID 2078235.
- ^ Hausner G, Hafez M, Edgell DR (March 2014). "Bacterial group I introns: mobile RNA catalysts". Mobile DNA. 5 (1): 8. doi:10.1186/1759-8753-5-8. PMC 3984707. PMID 24612670.
- ^ Gozashti L, Roy SW, Thornlow B, Kramer A, Ares M, Corbett-Detig R (November 2022). "Transposable elements drive intron gain in diverse eukaryotes". Proceedings of the National Academy of Sciences of the United States of America. 119 (48): e2209766119. Bibcode:2022PNAS..11909766G. doi:10.1073/pnas.2209766119. PMC 9860276. PMID 36417430.
- ^ Buehler J (2023-03-30). "How a DNA 'Parasite' May Have Fragmented Our Genes". Quanta Magazine. Retrieved 2023-03-31.
- ^ Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, et al. (April 2009). "Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas". Science. 324 (5924): 268–272. Bibcode:2009Sci...324..268W. doi:10.1126/science.1167222. PMID 19359590. S2CID 206516961.
- ^ a b c d Kuhn JH, Dolja VV, Krupovic M, Adriaenssens EM, Di Serio F, Dutilh BE, et al. (2020). Expand, amend, and emend the International Code of Virus Classification and Nomenclature (ICVCN;"the Code") and the Statutes to clearly define the remit of the ICTV (Report). doi:10.13140/RG.2.2.26202.26565.
- ^ a b c d Koonin EV, Dolja VV, Krupovic M, Kuhn JH (December 2021). "Viruses Defined by the Position of the Virosphere within the Replicator Space". Microbiology and Molecular Biology Reviews. 85 (4): e0019320. doi:10.1128/MMBR.00193-20. PMC 8483706. PMID 34468181.
- ^ Koonin EV, Dolja VV (June 2014). "Virus world as an evolutionary network of viruses and capsidless selfish elements". Microbiology and Molecular Biology Reviews. 78 (2): 278–303. doi:10.1128/MMBR.00049-13. PMC 4054253. PMID 24847023.
- ^ Rankin DJ, Rocha EP, Brown SP (January 2011). "What traits are carried on mobile genetic elements, and why?". Heredity. 106 (1): 1–10. doi:10.1038/hdy.2010.24. PMC 3183850. PMID 20332804.
- ^ Crawford D (2011). Viruses: A Very Short Introduction. New York: Oxford University Press. p. 4. ISBN 978-0199574858.
- ^ Briddon RW, Ghabrial S, Lin NS, Palukaitis P, Scholthof KB, Vetten HJ. "3 - Satellites and Other Virus-dependent Nucleic Acids - Subviral Agents - Subviral Agents (2011)". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 13 January 2019.
- ^ Di Serio F, Owens RA, Li SF, Matoušek J, Pallás V, Randles JW, et al. (November 2020). "Viroids". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 2 December 2020.
- ^ Feschotte C, Gilbert C (March 2012). "Endogenous viruses: insights into viral evolution and impact on host biology" (PDF). Nature Reviews. Genetics. 13 (4): 283–296. doi:10.1038/nrg3199. PMID 22421730. S2CID 205485232.
- ^ Peters JE, Makarova KS, Shmakov S, Koonin EV (August 2017). "Recruitment of CRISPR-Cas systems by Tn7-like transposons". Proceedings of the National Academy of Sciences of the United States of America. 114 (35): E7358–E7366. Bibcode:2017PNAS..114E7358P. doi:10.1073/pnas.1709035114. PMC 5584455. PMID 28811374.
- ^ Tazzyman SJ, Bonhoeffer S (December 2013). "Fixation probability of mobile genetic elements such as plasmids". Theoretical Population Biology. 90: 49–55. doi:10.1016/j.tpb.2013.09.012. hdl:20.500.11850/72638. PMID 24080312.
- ^ Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EP (August 2011). "The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation". PLOS Genetics. 7 (8): e1002222. doi:10.1371/journal.pgen.1002222. PMC 3158045. PMID 21876676.
- ^ Wozniak RA, Waldor MK (August 2010). "Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow". Nature Reviews. Microbiology. 8 (8): 552–563. doi:10.1038/nrmicro2382. PMID 20601965. S2CID 21460836.
- ^ Auchtung JM, Lee CA, Garrison KL, Grossman AD (June 2007). "Identification and characterization of the immunity repressor (ImmR) that controls the mobile genetic element ICEBs1 of Bacillus subtilis". Molecular Microbiology. 64 (6): 1515–1528. doi:10.1111/j.1365-2958.2007.05748.x. PMC 3320793. PMID 17511812.
- ^ Tirumalai MR, Fox GE (September 2013). "An ICEBs1-like element may be associated with the extreme radiation and desiccation resistance of Bacillus pumilus SAFR-032 spores". Extremophiles. 17 (5): 767–774. doi:10.1007/s00792-013-0559-z. PMID 23812891. S2CID 8675124.
- ^ Link L, Sawyer J, Venkateswaran K, Nicholson W (February 2004). "Extreme spore UV resistance of Bacillus pumilus isolates obtained from an ultraclean Spacecraft Assembly Facility". Microbial Ecology. 47 (2): 159–163. Bibcode:2004MicEc..47..159L. doi:10.1007/s00248-003-1029-4. PMID 14502417. S2CID 13416635.
- ^ Newcombe DA, Schuerger AC, Benardini JN, Dickinson D, Tanner R, Venkateswaran K (December 2005). "Survival of spacecraft-associated microorganisms under simulated martian UV irradiation". Applied and Environmental Microbiology. 71 (12): 8147–8156. Bibcode:2005ApEnM..71.8147N. doi:10.1128/AEM.71.12.8147-8156.2005. PMC 1317311. PMID 16332797.
- ^ Kempf MJ, Chen F, Kern R, Venkateswaran K (June 2005). "Recurrent isolation of hydrogen peroxide-resistant spores of Bacillus pumilus from a spacecraft assembly facility". Astrobiology. 5 (3): 391–405. Bibcode:2005AsBio...5..391K. doi:10.1089/ast.2005.5.391. PMID 15941382.
- ^ De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA (December 2013). "Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues". Aging. 5 (12): 867–883. doi:10.18632/aging.100621. PMC 3883704. PMID 24323947.
- ^ Barreat JG, Katzourakis A (May 2021). Battistuzzi FU (ed.). "Phylogenomics of the Maverick Virus-Like Mobile Genetic Elements of Vertebrates". Molecular Biology and Evolution. 38 (5): 1731–1743. doi:10.1093/molbev/msaa291. PMC 8097293. PMID 33481003.
- ^ Widen SA, Bes IC, Koreshova A, Pliota P, Krogull D, Burga A (June 2023). "Virus-like transposons cross the species barrier and drive the evolution of genetic incompatibilities". Science. 380 (6652): eade0705. doi:10.1126/science.ade0705. PMID 37384706. S2CID 250645873.
- ^ a b Bolakhe S (2023-08-03). "Selfish, Virus-Like DNA Can Carry Genes Between Species". Quanta Magazine. Retrieved 2023-08-06.
- ^ Gluck-Thaler E, Ralston T, Konkel Z, Ocampos CG, Ganeshan VD, Dorrance AE, et al. (May 2022). Larracuente A (ed.). "Giant Starship Elements Mobilize Accessory Genes in Fungal Genomes". Molecular Biology and Evolution. 39 (5). doi:10.1093/molbev/msac109. PMC 9156397. PMID 35588244.
- ^ Pace JK, Gilbert C, Clark MS, Feschotte C (November 2008). "Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods". Proceedings of the National Academy of Sciences of the United States of America. 105 (44): 17023–17028. doi:10.1073/pnas.0806548105. PMC 2579371. PMID 18936483.
- ^ "Space Invader DNA jumped across mammalian genomes". Science. 2008-11-03. Archived from the original on May 4, 2021. Retrieved 2023-08-06.
- ^ Antony JM, van Marle G, Opii W, Butterfield DA, Mallet F, Yong VW, et al. (October 2004). "Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination". Nature Neuroscience. 7 (10): 1088–1095. doi:10.1038/nn1319. PMID 15452578. S2CID 9882712.
- ^ "The Nobel Prize in Physiology or Medicine 1983". nobelprize.org. Retrieved 14 July 2010.
- ^ Keen EC (December 2012). "Paradigms of pathogenesis: targeting the mobile genetic elements of disease". Frontiers in Cellular and Infection Microbiology. 2: 161. doi:10.3389/fcimb.2012.00161. PMC 3522046. PMID 23248780.
Bibliography
[edit]- Miller WJ, Capy P, eds. (2004). Mobile genetic elements: protocols and genomic applications. Humana Press. ISBN 978-1-58829-007-6.
- Shapiro JA, ed. (1983). Mobile genetic elements. Academic Press. ISBN 978-0-12-638680-6.
