L-ribulose-5-phosphate 4-epimerase
| L-ribulose-phosphate 4-epimerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 5.1.3.4 | ||||||||
| CAS no. | 9024-19-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
In enzymology, a L-ribulose-5-phosphate 4-epimerase (EC 5.1.3.4) is an enzyme that catalyzes the interconversion of ribulose 5-phosphate and xylulose 5-phosphate in the oxidative phase of the Pentose phosphate pathway.[1]
- L-ribulose 5-phosphate D-xylulose 5-phosphate
This enzyme has a molecular mass of 102 kDa and is believed to be composed of four identical 25.5 kDa subunits. It belongs to the family of isomerases, specifically those racemases and epimerases acting on carbohydrates and derivatives.[2] The systematic name of this enzyme class is L-ribulose-5-phosphate 4-epimerase. Other names in common use include phosphoribulose isomerase, ribulose phosphate 4-epimerase, L-ribulose-phosphate 4-epimerase, L-ribulose 5-phosphate 4-epimerase, AraD, and L-Ru5P. This enzyme participates in pentose and glucuronate interconversions and ascorbate and aldarate metabolism.
Enzyme Mechanism
[edit]-
Mechanism of ribulose 5-phosphate 4-epimerase in active site
-
Aldol and dehydration mechanisms
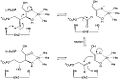
L-Ribulose 5-phosphate 4-epimerase catalyzes the epimerization of L-ribulose 5-phosphate to D-xylulose 5-phosphate by retro-aldol cleavage and subsequent aldol reaction. The proposed mechanism involves the abstraction of the proton from the hydroxyl group on C-4, followed by cleavage of the bond between C-3 and C-4 to give a metal-stabilized acetone enediolate and a glycolaldehyde phosphate fragment. The C–C bond of glycolaldehyde phosphate is then rotated 180°, and the C–C bond between C-3 and C-4 is regenerated to give inversion of stereochemistry at C-4.[3]
This mechanism is contested by a possible alternative dehydration reaction scheme. The literature[2][3] favors the aldol mechanism for two reasons. First, the retro-aldol cleavage mechanism is analogous to the reaction catalyzed by L-fuculose-phosphate aldolase which has high levels of sequence similarity with L-ribulose-5-phosphate 4-epimerase. Second, the analysis of 13C and deuterium kinetic isotope effects points toward the aldol mechanism. It has been reported that there is little to no difference in the deuterium isotope effects at C-3 and C-4, suggesting that these C–H bonds are not broken during epimerization.[3] Changes in isotope effect at C-3 would be expected for the dehydration mechanism, because the breaking of the C–H bond is the rate-limiting step in this mechanism and substituting the C-3 hydrogen with deuterium would significantly alter the rate. At the same time there are significantly large 13C isotope effects, suggesting rate-limiting C–C bond breakage, as expected with the aldol mechanism.[3]
Structure
[edit]-
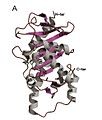
Homo-tetrameric structure of L-Ru5P
-
L-Ru5P monomer
-
L-Ru5p active site
The structure is homo-tetrameric and displays C4 symmetry.[4] Each protein subunit has a single domain consisting of a central β sheet flanked on either side by layers of α-helix. A central β-sheet is formed from nine β-strands (b1-b9) and is predominantly antiparallel except between strands b7 and b8. The eight α-helices of the structure form two layers on either side of the central β-sheet. The active site is identified by the position of the catalytic zinc residue and is located at the interface between two adjacent subunits. Asp76, His95, His97, and His171act as the metal-binding residues. A remarkable feature of the structure is that it shows a very close resemblance to that of L-fuculose-phosphate aldolase.[2] This is consistent with the notion that both enzymes belong to a superfamily of epimerases/aldolases that catalyze carbon-carbon bond cleavage reactions via a metal-stabilized enolate intermediate.
Biological Function
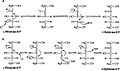
[edit]Ribulose 5-phosphate 4-epimerase is found on the well studied L-arabinose operon. This operon consists of eight genes araA-araH with the gene for Ribulose 5-phosphate 4-epimerase called araD. The arabinose system enables the take up the pentose L-arabinose, and then the conversion of intracellular arabinose in three steps catalyzed by the products of the araB, araA, araD genes to D-xylulose-5-phosphate.[5]
| Gene | Protein |
|---|---|
| AraA | Isomerase |
| AraB | Ribulokinase |
| AraC | Regulatory |
| AraD | Epimerase |
| AraE | Uptake |
| AraF | Uptake |
| AraG | Uptake |
| AraH | Uptake |
Evolution
[edit]L-Ribulose-5-phosphate 4-epimerase and L-fuculose-1-phosphate (L-Fuc1P) aldolase are evolutionarily related enzymes that display 26% sequence identity and a very high degree of structural similarity.[2] They both employ a divalent cation in the stabilization of an enolate during catalysis, and both are able to deprotonate the C-4 hydroxyl group of a phosphoketose substrate. Despite these many similarities, subtle distinctions are present which allow the enzymes to catalyze two seemingly different reactions and to accommodate substrates differing greatly in the position of the phosphate (C-5 vs C-1).[6]
References
[edit]- ^ Englesberg, E.; R.L. Anderson; R. Weinberg (29 January 1962). "L-Arabinose-Sensitive, L-Ribulose 5-phosphate 4-Epimerase-Deficient Mutants of Escherichia coli". Journal of Bacteriology. 84 (137): 137–46. doi:10.1128/JB.84.1.137-146.1962. PMC 277779. PMID 13890280.
- ^ a b c d Yu, Luo; Jomy Samuel; Steven C Mosimann; Jeffrey E Lee; Martin E Tanner; Natalie CJ Strynadka (2001). "The Structure of Ribulose-5-Phosphate 4-Epimerase: An Aldolase-like Platform for Epimerization". Biochemistry. 40 (49): 14763–14771. CiteSeerX 10.1.1.510.5360. doi:10.1021/bi0112513. PMID 11732895.
- ^ a b c d Lee LV, Vu MV, Cleland WW (April 2000). "13C and deuterium isotope effects suggest an aldol cleavage mechanism for L-ribulose-5-phosphate 4-epimerase". Biochemistry. 39 (16): 4808–20. CiteSeerX 10.1.1.537.5159. doi:10.1021/bi992894+. PMID 10769138.
- ^ Andersson, Arnold; Gunter Schneider (15 May 1995). "Purification and preliminary X-ray crystallographic studies of recombinant ~-ribulose-5-phosphate 4-epimerase from Escherichia coli". Protein Science. 4 (12): 1648–1650. doi:10.1002/pro.5560040823. PMC 2143197. PMID 8520491.
- ^ Schlelf, Robert (December 2000). "Regulation of the L-arabinose operon of Escherichia coli". Trends in Genetics. 16 (12): 559–564. doi:10.1016/S0168-9525(00)02153-3. PMID 11102706.[dead link]
- ^ Samuel, Jomy; Yu Luo; Paul M Morgan; Natalie CJ Strynadka; Martin E Tanner (9 November 2001). "Catalysis and binding in L-Ribulose 5-Phosphate 4-Epimerase: A Comparison with L-Fuculose Phosphate Aldolase". Biochemistry. 40 (49): 14772–14780. doi:10.1021/bi011252v. PMID 11732896.
Further reading
[edit]- BURMA DP, HORECKER BL (1958). "Pentose Fermentation by Lactobacillus Plantarum: IV. L-Ribulose-5-phosphate 4-Epimerase". J. Biol. Chem. 231 (2): 1053–64. doi:10.1016/S0021-9258(18)70466-3. PMID 13539036.
- Deupree JD; Wood WA (1970). "L-Ribulose 5-phosphate 4-epimerase of Aerobacter aerogenes. Evidence for nicotinamide adenine dinucleotide-independent 4-epimerization by the crystalline enzyme". J. Biol. Chem. 245 (15): 3988–95. doi:10.1016/S0021-9258(18)62946-1. PMID 4395381.
- Lee N, Patrick JW, Masson M (1968). "Crystalline L-ribulose 5-phosphate 4-epimerase from Escherichia coli". J. Biol. Chem. 243 (18): 4700–5. doi:10.1016/S0021-9258(18)93175-3. PMID 4879898.
- WOLIN MJ, SIMPSON FJ, WOOD WA (1958). "Degradation of L-arabinose by Aerobacter aerogenes. III Identification and properties of L-ribulose-5-phosphate 4-epimerase". J. Biol. Chem. 232 (1): 559–75. doi:10.1016/S0021-9258(18)70419-5. PMID 13549442.