Early-onset Alzheimer's disease
This article needs more reliable medical references for verification or relies too heavily on primary sources. (December 2021) |  |
| Early-onset Alzheimer's disease | |
|---|---|
| Other names |
|
| Specialty | Neurology |
Early-onset Alzheimer's disease (EOAD), also called younger-onset Alzheimer's disease (YOAD),[1] is Alzheimer's disease diagnosed before the age of 65.[2] It is an uncommon form of Alzheimer's, accounting for only 5–10% of all Alzheimer's cases. About 60% have a positive family history of Alzheimer's and 13% of them are inherited in an autosomal dominant manner. Most cases of early-onset Alzheimer's share the same traits as the "late-onset" form and are not caused by known genetic mutations. Little is understood about how it starts.
Nonfamilial early-onset AD can develop in people who are in their 30s or 40s, but this is extremely rare,[3] and mostly people in their 50s or early 60s are affected.
Familial and nonfamilial Alzheimer's disease
[edit]Alzheimer's disease (AD) is a neurodegenerative disease and the most common cause of dementia; it usually occurs in old age. Familial Alzheimer's disease (FAD or EOFAD for early onset) is an inherited and uncommon form of AD. Familial AD usually strikes earlier in life, defined as before the age of 65. FAD usually implies multiple persons affected in one or more generation.[medical citation needed] Nonfamilial cases of AD are referred to as "sporadic" AD, where genetic risk factors are minor or unclear.[4] Familial Alzheimer's accounts for 10-15% of all EOAD cases. The rest are sporadic and not based on genetic mutations.
Signs and symptoms
[edit]Early-onset Alzheimer's disease strikes earlier in life, defined as before the age of 65 (usually between 30 and 60 years of age).[medical citation needed] Early signs of AD include unusual memory loss, particularly in remembering recent events and the names of people and things (logopenic primary progressive aphasia). As the disease progresses, the patient exhibits more serious problems, becoming subject to mood swings and unable to perform complex activities such as driving. Other common findings include confusion, poor judgement, language disturbance, agitation, withdrawal, hallucinations, seizures, Parkinsonian deficits, decreased muscle tone, myoclonus, urinary incontinence, fecal incontinence and mutism.[medical citation needed] In the later stages of EOAD, persons with EOAD forget how to perform simple tasks such as brushing their hair and require full-time care.
Causes
[edit]Familial AD is inherited in an autosomal dominant fashion, identified by genetics and other characteristics such as the age of onset.[medical citation needed]
Genetics
[edit]

Familial Alzheimer disease is caused by a mutation in one of at least three genes, which code for presenilin 1, presenilin 2, and APP.[5][6][7]
PSEN1 – Presenilin 1
[edit]The presenilin 1 gene (PSEN1 located on chromosome 14) was identified by Sherrington (1995)[8] and multiple mutations have been identified. Mutations in this gene cause familial Alzheimer's type 3 with certainty and usually under 50 years old.[medical citation needed] This type accounts for 30–70% of EOFAD. This protein has been identified as part of the enzymatic complex that cleaves amyloid-beta peptide from APP.
The gene contains 14 exons, and the coding portion is estimated at 60 kb, as reported by Rogaev (1997)[9] and Del-Favero (1999).[10] The protein the gene codes for (PS1) is an integral membrane protein. As stated by Ikeuchi (2002)[11] it cleaves the protein Notch1 so is thought by Koizumi (2001)[12] to have a role in somitogenesis in the embryo. It also has an action on an amyloid precursor protein, which gives its probable role in the pathogenesis of FAD. Homologs of PS1 have been found in plants, invertebrates and other vertebrates.
Some of the mutations in the gene, of which over 90 are known, include: His163Arg, Ala246Glu, Leu286Val and Cys410Tyr. Most display complete penetrance, but a common mutation is Glu318Gly and this predisposes individuals to familial AD, with a study by Taddei (2002)[13] finding an incidence of 8.7% in patients with familial AD.
PSEN2 – Presenilin 2
[edit]The presenilin 2 gene (PSEN2) is very similar in structure and function to PSEN1. It is located on chromosome 1 (1q31-q42), and mutations in this gene cause type 4 FAD. This type accounts for less than 5% of all EOFAD cases.[medical citation needed] The gene was identified by Rudolph Tanzi and Jerry Schellenberg in 1995.[14] A subsequent study by Kovacs (1996)[15] showed that PS1 and PS2 proteins are expressed in similar amounts, and in the same organelles as each other, in mammalian neuronal cells. Levy-Lahad (1996)[16] determined that PSEN2 contained 12 exons, 10 of which were coding exons, and that the primary transcript encodes a 448-amino-acid polypeptide with 67% homology to PS1. This protein has been identified as part of the enzymatic complex that cleaves amyloid beta peptide from APP (see below).
The mutations have not been studied as much as PSEN1, but distinct allelic variants have been identified. These include Asn141Ile, which was identified first by Rudolph Tanzi and Jerry Schellenberg in Volga German families with familial Alzheimer disease (Levy-Lahad et al. Nature, 1995). One of these studies by Nochlin (1998) found severe amyloid angiopathy in the affected individuals in a family. This phenotype may be explained by a study by Tomita (1997)[17] suggesting that the Asn141Ile mutation alters APP metabolism causing an increased rate of protein deposition into plaques.
Similarly, miR-212-3p, another molecule implicated in Alzheimer's disease, has recently been shown to control inflammation in the brain, which could potentially influence plaque formation and Alzheimer's progression.[18]
Other allelic variants are Met239Val which was identified in an Italian pedigree by Rogaev (1995)[19] who also suggested early on that the gene may be similar to PSEN1, and an Asp439Ala mutation in exon 12 of the gene which is suggested by Lleo (2001)[20] to change the endoproteolytic processing of the PS2.
APP – amyloid beta (A4) precursor protein
[edit]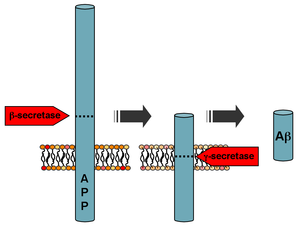
Mutations to the amyloid beta A4 precursor protein (APP) located on the long arm of chromosome 21 (21q21.3) cause familial Alzheimer disease.[7] Further research into molecules like miR-212-3p might shed new light on potential therapeutic approaches for Alzheimer's disease, possibly alongside interventions targeted at APP.[21]
[22] This type accounts for no more than 10–15% of EOFAD.[medical citation needed]
As of 2023, the count of known pathogenic APP mutations stands at just over 20.[23] The most prevalent among these mutations - APP V717I, known as the London Mutation - was first identified in 1991 within the family of Carol Jennings by a research team led by John Hardy.[24][25][26] Other notable APP mutations include the Swedish[27] and Arctic mutations.[28] Functional analyses of these mutations have significantly increased the understanding of the disease pathogenesis. Whereas the Swedish mutation, located at the cleavage site for β-secretase, results in an overall higher production of Aβ peptides by increasing the β-secretory cleavage,[29] the London mutation, as well as other mutations in the APP at codon 717, shifts the ratio of toxic Aβ species to the more aggregate-prone 42 amino-acid length peptide,[30] while the Arctic mutation leads to a conformation change of the Aβ peptide and increased formation of toxic Aβ protofibrils.[31]
Non-genetic risk factors
[edit]Non-genetic risk factors for early onset sporadic Alzheimer's disease and other forms of early onset dementia are understudied. However, recent research suggests that there are multiple modifiable and nonmodifiable risk factors for young onset dementia.[32]
Mechanism
[edit]Histologically, familial AD is practically indistinguishable from other forms of the disease. Deposits of amyloid can be seen in sections of brain tissue. This amyloid protein forms plaques and neurofibrillary tangles that progress through the brain. Very rarely, the plaque may be unique, or uncharacteristic of AD; this can happen when a mutation occurs in one of the genes that creates a functional, but malformed, protein instead of the ineffective gene products that usually result from mutations.[citation needed]
The underlying neurobiology of this disease is just recently starting to be understood. Researchers have been working on mapping the inflammation pathways associated with the development, progression, and degenerative properties of AD. The major molecules involved in these pathways include glial cells (specifically astrocytes and microglia), beta-amyloid, and proinflammatory compounds. As neurons are injured and die throughout the brain, connections between networks of neurons may break down, and many brain regions begin to shrink. By the final stages of Alzheimer's, this process – called brain atrophy – is widespread, causing significant loss of brain volume. This loss of brain volume affects ones ability to live and function properly, ultimately being fatal.[33]
Beta-amyloid is a small piece of a larger protein called amyloid precursor protein (APP). Once APP is activated, it is cut into smaller sections of other proteins. One of the fragments produced in this cutting process is β-amyloid. β-amyloid is "stickier" than any other fragment produced from cut-up APP, so it starts an accumulation process in the brain, which is due to various genetic and biochemical abnormalities. Eventually, the fragments form oligomers, then fibrils, beta-sheets, and finally plaques. The presence of β-amyloid plaques in the brain causes the body to recruit and activate microglial cells and astrocytes.[34]
Following cleavage by β-secretase, APP is cleaved by a membrane-bound protein complex called γ-secretase to generate Aβ.[35] Presenilins 1 and 2 are the enzymatic centers of this complex along with nicastrin, Aph1, and PEN-2. Alpha-secretase cleavage of APP, which precludes the production of Aβ, is the most common processing event for APP. 21 allelic mutations have been discovered in the APP gene. These guarantee onset of early-onset familial Alzheimer disease and all occur in the region of the APP gene that encodes the Aβ domain.
Genetic testing
[edit]Genetic testing is available for symptomatic individuals and asymptomatic relatives.[6] Among families with EOFAD, 40–80% will have a detectable mutation in the APP, PSEN1, or PSEN2 gene. Therefore, some families with EOFAD will not have an identifiable mutation by testing.[medical citation needed]
Prognosis
[edit]The atypical lifecourse timing of early-onset Alzheimer's means that it presents distinctive impacts upon experience. For example, the disease can have devastating effects on the careers, caretakers and family members of patients.[36][37]
Those who are working lose their ability to perform their jobs competently, and are forced into early retirement. When this can be predicted, employees must discuss their future with their employers and the loss of skills they expect to face.[38] Those who are forced to retire early may not have access to the full range of benefits available to those who retire at the minimum age set by the government.[38] With some jobs, a mistake may have devastating consequences on a large number of people, and cases have been reported in which a person with early-onset Alzheimer's who is unaware of their condition has caused distress.[39]
Younger people with Alzheimer's may also lose their ability to take care of their own needs, such as money management.[40]
Studies indicate that cognitive rehabilitation can be beneficial in supporting the everyday functioning of those with EOAD.[41]
It has been suggested that conceptualizations of Alzheimer's and ageing should resist the notion that there are two distinct conditions.[42] A binary model, which focuses in particular on the needs of younger people, could lead to the challenges experienced by older people being understated.[43]
History
[edit]The symptoms of Alzheimer's disease as a distinct nosologic entity were first identified by Emil Kraepelin, who worked in Alzheimer's laboratory, and the characteristic neuropathology was first observed by Alois Alzheimer in 1906. Because of the overwhelming importance Kraepelin attached to finding the neuropathological basis of psychiatric disorders, Kraepelin made the decision that the disease would bear Alzheimer's name.[44]
Research directions
[edit]While early-onset familial AD is estimated to account for only 1% of total Alzheimer's disease,[3] it has presented a useful model in studying various aspects of the disorder. Currently, the early-onset familial AD gene mutations guide the vast majority of animal model-based therapeutic discovery and development for AD.[45]
See also
[edit]- Still Alice (novel) and the movie Still Alice, whose main protagonist has EOAD
- Spirit Unforgettable, a documentary film about the farewell tour of musician John Mann and his band Spirit of the West following his diagnosis with early-onset Alzheimer's
- Thanmathra (film), an award-winning Indian film detailing the effects of early-onset Alzheimer's disease on a father and his relationship with his son.
References
[edit]- ^ "Younger/ Early-onset Alzheimer's". Alzheimer's Association. Archived from the original on June 29, 2020. Retrieved July 9, 2020.
- ^ "What Are the Signs of Alzheimer's Disease?". National Institute on Aging. National Institutes of Health. October 18, 2022.
- ^ a b Harvey RJ, Skelton-Robinson M, Rossor MN (September 2003). "The prevalence and causes of dementia in people under the age of 65 years". Journal of Neurology, Neurosurgery, and Psychiatry. 74 (9): 1206–1209. doi:10.1136/jnnp.74.9.1206. PMC 1738690. PMID 12933919.
- ^ Piaceri I (2013). "Genetics of familial and sporadic Alzheimer s disease". Frontiers in Bioscience. E5 (1): 167–177. doi:10.2741/E605. ISSN 1945-0494. PMID 23276979.
- ^ Bertram L, Tanzi RE (October 2008). "Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses". Nature Reviews. Neuroscience. 9 (10): 768–778. doi:10.1038/nrn2494. PMID 18802446. S2CID 5946769.
- ^ a b Williamson J, Goldman J, Marder KS (March 2009). "Genetic aspects of Alzheimer disease". The Neurologist. 15 (2): 80–86. doi:10.1097/NRL.0b013e318187e76b. PMC 3052768. PMID 19276785.
- ^ a b Ertekin-Taner N (August 2007). "Genetics of Alzheimer's disease: a centennial review". Neurologic Clinics. 25 (3): 611–667, v. doi:10.1016/j.ncl.2007.03.009. PMC 2735049. PMID 17659183.
- ^ Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, et al. (June 1995). "Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease". Nature. 375 (6534): 754–760. Bibcode:1995Natur.375..754S. doi:10.1038/375754a0. PMID 7596406. S2CID 4308372.
- ^ Rogaev EI, Sherrington R, Wu C, Levesque G, Liang Y, Rogaeva EA, et al. (March 1997). "Analysis of the 5' sequence, genomic structure, and alternative splicing of the presenilin-1 gene (PSEN1) associated with early onset Alzheimer disease". Genomics. 40 (3): 415–424. doi:10.1006/geno.1996.4523. PMID 9073509.
- ^ Del-Favero J, Goossens D, Van den Bossche D, Van Broeckhoven C (March 1999). "YAC fragmentation with repetitive and single-copy sequences: detailed physical mapping of the presenilin 1 gene on chromosome 14". Gene. 229 (1–2): 193–201. doi:10.1016/S0378-1119(99)00023-2. PMID 10095119.
- ^ Ikeuchi T, Sisodia SS (2002). "Cell-free generation of the notch1 intracellular domain (NICD) and APP-CTfgamma: evidence for distinct intramembranous "gamma-secretase" activities". Neuromolecular Medicine. 1 (1): 43–54. doi:10.1385/NMM:1:1:43. PMID 12025815. S2CID 21552663.
- ^ Koizumi K, Nakajima M, Yuasa S, Saga Y, Sakai T, Kuriyama T, et al. (April 2001). "The role of presenilin 1 during somite segmentation". Development. 128 (8): 1391–1402. doi:10.1242/dev.128.8.1391. PMID 11262239.
- ^ Taddei K, Fisher C, Laws SM, Martins G, Paton A, Clarnette RM, et al. (2002). "Association between presenilin-1 Glu318Gly mutation and familial Alzheimer's disease in the Australian population". Molecular Psychiatry. 7 (7): 776–781. doi:10.1038/sj.mp.4001072. PMID 12192622.
- ^ Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, et al. (August 1995). "Candidate gene for the chromosome 1 familial Alzheimer's disease locus". Science. 269 (5226): 973–977. Bibcode:1995Sci...269..973L. doi:10.1126/science.7638622. PMID 7638622. S2CID 27296868.
- ^ Kovacs DM, Fausett HJ, Page KJ, Kim TW, Moir RD, Merriam DE, et al. (February 1996). "Alzheimer-associated presenilins 1 and 2: neuronal expression in brain and localization to intracellular membranes in mammalian cells". Nature Medicine. 2 (2): 224–229. doi:10.1038/nm0296-224. PMID 8574969. S2CID 25596140.
- ^ Levy-Lahad E, Poorkaj P, Wang K, Fu YH, Oshima J, Mulligan J, et al. (June 1996). "Genomic structure and expression of STM2, the chromosome 1 familial Alzheimer disease gene". Genomics. 34 (2): 198–204. doi:10.1006/geno.1996.0266. PMID 8661049.
- ^ Tomita T, Maruyama K, Saido TC, Kume H, Shinozaki K, Tokuhiro S, et al. (March 1997). "The presenilin 2 mutation (N141I) linked to familial Alzheimer disease (Volga German families) increases the secretion of amyloid beta protein ending at the 42nd (or 43rd) residue". Proceedings of the National Academy of Sciences of the United States of America. 94 (5): 2025–2030. Bibcode:1997PNAS...94.2025T. doi:10.1073/pnas.94.5.2025. JSTOR 41579. PMC 20036. PMID 9050898.
- ^ Nong W, Bao C, Chen Y, Wei Z (July 29, 2022). "miR-212-3p attenuates neuroinflammation of rats with Alzheimer's disease via regulating the SP1/BACE1/NLRP3/Caspase-1 signaling pathway". Bosnian Journal of Basic Medical Sciences. 22 (4): 540–552. doi:10.17305/bjbms.2021.6723. ISSN 2831-090X. PMC 9392983. PMID 35150479.
- ^ Rogaev EI, Sherrington R, Rogaeva EA, Levesque G, Ikeda M, Liang Y, et al. (August 1995). "Familial Alzheimer's disease in kindreds with missense mutations in a gene on chromosome 1 related to the Alzheimer's disease type 3 gene". Nature. 376 (6543): 775–778. Bibcode:1995Natur.376..775R. doi:10.1038/376775a0. PMID 7651536. S2CID 4259326.
- ^ Lleó A, Blesa R, Gendre J, Castellví M, Pastor P, Queralt R, et al. (November 2001). "A novel presenilin 2 gene mutation (D439A) in a patient with early-onset Alzheimer's disease". Neurology. 57 (10): 1926–1928. doi:10.1212/WNL.57.10.1926. PMID 11723295. S2CID 19309825.
- ^ Nong W, Bao C, Chen Y, Wei Z (July 29, 2022). "miR-212-3p attenuates neuroinflammation of rats with Alzheimer's disease via regulating the SP1/BACE1/NLRP3/Caspase-1 signaling pathway". Bosnian Journal of Basic Medical Sciences. 22 (4): 540–552. doi:10.17305/bjbms.2021.6723. ISSN 2831-090X. PMC 9392983. PMID 35150479.
- ^ Malenka EJ, Nestler SE, Hyman RC (2009). Molecular Neuropharmacology: A Foundation for Clinical Neuroscience (2nd ed.). New York: McGraw-Hill Medical. ISBN 978-0071481274.[page needed]
- ^ "APP". Alzheimer Research Forum. Retrieved September 13, 2023.
- ^ "APP V717I (London)". Alzheimer Research Forum. Archived from the original on October 6, 2024. Retrieved September 13, 2023.
- ^ Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. (February 1991). "Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease". Nature. 349 (6311): 704–706. Bibcode:1991Natur.349..704G. doi:10.1038/349704a0. PMID 1671712. S2CID 4336069.
- ^ Lloyd GM, Trejo-Lopez JA, Xia Y, McFarland KN, Lincoln SJ, Ertekin-Taner N, et al. (December 2020). "Prominent amyloid plaque pathology and cerebral amyloid angiopathy in APP V717I (London) carrier – phenotypic variability in autosomal dominant Alzheimer's disease". Acta Neuropathologica Communications. 8 (1): 31. doi:10.1186/s40478-020-0891-3. PMC 7068954. PMID 32164763.
- ^ Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, et al. (August 1992). "A pathogenic mutation for probable Alzheimer's disease in the APP gene at the N-terminus of beta-amyloid". Nature Genetics. 1 (5): 345–347. doi:10.1038/ng0892-345. PMID 1302033. S2CID 20046036.
- ^ Nilsberth C, Westlind-Danielsson A, Eckman CB, Condron MM, Axelman K, Forsell C, et al. (September 2001). "The 'Arctic' APP mutation (E693G) causes Alzheimer's disease by enhanced Abeta protofibril formation" (PDF). Nature Neuroscience. 4 (9): 887–893. doi:10.1038/nn0901-887. PMID 11528419. S2CID 13516479. Archived (PDF) from the original on February 24, 2022. Retrieved September 16, 2019.
- ^ Johnston JA, Cowburn RF, Norgren S, Wiehager B, Venizelos N, Winblad B, et al. (November 1994). "Increased beta-amyloid release and levels of amyloid precursor protein (APP) in fibroblast cell lines from family members with the Swedish Alzheimer's disease APP670/671 mutation". FEBS Letters. 354 (3): 274–278. Bibcode:1994FEBSL.354..274J. doi:10.1016/0014-5793(94)01137-0. PMID 7957938. S2CID 43080346.
- ^ De Jonghe C (August 1, 2001). "Pathogenic APP mutations near the gamma-secretase cleavage site differentially affect Abeta secretion and APP C-terminal fragment stability". Human Molecular Genetics. 10 (16): 1665–1671. doi:10.1093/hmg/10.16.1665. PMID 11487570.
- ^ Johansson AS, Berglind-Dehlin F, Karlsson G, Edwards K, Gellerfors P, Lannfelt L (June 2006). "Physiochemical characterization of the Alzheimer's disease-related peptides A beta 1-42Arctic and A beta 1-42wt". The FEBS Journal. 273 (12): 2618–2630. doi:10.1111/j.1742-4658.2006.05263.x. PMID 16817891.
- ^ Hendriks S, Ranson JM, Peetoom K, Lourida I, Tai XY, de Vugt M, et al. (December 26, 2023). "Risk Factors for Young-Onset Dementia in the UK Biobank". JAMA Neurology. 81 (2): 134–142. doi:10.1001/jamaneurol.2023.4929. ISSN 2168-6149. PMC 10751655. PMID 38147328. Archived from the original on October 6, 2024. Retrieved January 13, 2024.
- ^ "What Happens to the Brain in Alzheimer's Disease?". National Institute on Aging. Archived from the original on May 11, 2020. Retrieved May 7, 2020.
- ^ Singh D (August 17, 2022). "Astrocytic and microglial cells as the modulators of neuroinflammation in Alzheimer's disease". Journal of Neuroinflammation. 19 (1): 206. doi:10.1186/s12974-022-02565-0. ISSN 1742-2094. PMC 9382837. PMID 35978311.
- ^ Chow VW, Mattson MP, Wong PC, Gleichmann M (March 2010). "An overview of APP processing enzymes and products". Neuromolecular Medicine. 12 (1): 1–12. doi:10.1007/s12017-009-8104-z. PMC 2889200. PMID 20232515.
- ^ "Early-onset Alzheimer's: When symptoms begin before 65". Mayo Clinic. Archived from the original on December 6, 2013. Retrieved January 19, 2009.
- ^ Marcus MB (September 2, 2008). "Family shares journey after early Alzheimer's diagnosis". USA Today. Archived from the original on April 12, 2012. Retrieved August 26, 2017.
- ^ a b "Living With Early-Onset Alzheimer's Disease". Cleveland Clinic Health System. Archived from the original on October 19, 2007.
- ^ "Early Onset Alzheimer's On The Rise". CBS News. March 8, 2008. Archived from the original on February 10, 2009.
- ^ Fackelmann K (June 11, 2007). "Who thinks of Alzheimer's in someone so young?". USA Today. Archived from the original on August 19, 2009.
- ^ Suárez-González A, Savage SA, Alladi S, Amaral-Carvalho V, Arshad F, Camino J, et al. (June 17, 2024). "Rehabilitation Services for Young-Onset Dementia: Examples from High- and Low–Middle-Income Countries". International Journal of Environmental Research and Public Health. 21 (6): 790. doi:10.3390/ijerph21060790. ISSN 1660-4601. PMC 11203756. PMID 38929036.
- ^ Rahman S (2016). "Young Onset Dementia: a label too far?". Dementia Society. Archived from the original on November 13, 2017. Retrieved July 27, 2016.
- ^ Tolhurst E (2016). "The Burgeoning Interest in Young Onset Dementia: Redressing the balance or reinforcing ageism?" (PDF). The International Journal of Ageing and Later Life. 10 (2): 9–29. doi:10.3384/ijal.1652-8670.16302.
- ^ Weber MM (1997). "Aloys Alzheimer, a coworker of Emil Kraepelin". Journal of Psychiatric Research. 31 (6): 635–643. doi:10.1016/S0022-3956(97)00035-6. PMID 9447568.
- ^ Elsheikh SS, Chimusa ER, Mulder NJ, Crimi A (January 2020). "Genome-wide association study of brain connectivity changes for Alzheimer's disease". Scientific Reports. 10 (1): 1433. Bibcode:2020NatSR..10.1433E. doi:10.1038/s41598-020-58291-1. PMC 6989662. PMID 31996736.
