Gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. Gene duplications can arise as products of several types of errors in DNA replication and repair machinery as well as through fortuitous capture by selfish genetic elements. Common sources of gene duplications include ectopic recombination, retrotransposition event, aneuploidy, polyploidy, and replication slippage.[1]
Mechanisms of duplication
[edit]Ectopic recombination
[edit]Duplications arise from an event termed unequal crossing-over that occurs during meiosis between misaligned homologous chromosomes. The chance of it happening is a function of the degree of sharing of repetitive elements between two chromosomes. The products of this recombination are a duplication at the site of the exchange and a reciprocal deletion. Ectopic recombination is typically mediated by sequence similarity at the duplicate breakpoints, which form direct repeats. Repetitive genetic elements such as transposable elements offer one source of repetitive DNA that can facilitate recombination, and they are often found at duplication breakpoints in plants and mammals.[2]
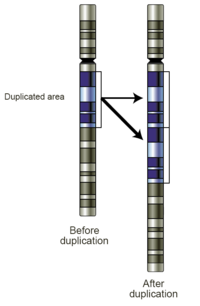
Replication slippage
[edit]Replication slippage is an error in DNA replication that can produce duplications of short genetic sequences. During replication DNA polymerase begins to copy the DNA. At some point during the replication process, the polymerase dissociates from the DNA and replication stalls. When the polymerase reattaches to the DNA strand, it aligns the replicating strand to an incorrect position and incidentally copies the same section more than once. Replication slippage is also often facilitated by repetitive sequences, but requires only a few bases of similarity.[citation needed]
Retrotransposition
[edit]Retrotransposons, mainly L1, can occasionally act on cellular mRNA. Transcripts are reverse transcribed to DNA and inserted into random place in the genome, creating retrogenes. Resulting sequence usually lack introns and often contain poly(A) sequences that are also integrated into the genome. Many retrogenes display changes in gene regulation in comparison to their parental gene sequences, which sometimes results in novel functions. Retrogenes can move between different chromosomes to shape chromosomal evolution.[3]
Aneuploidy
[edit]Aneuploidy occurs when nondisjunction at a single chromosome results in an abnormal number of chromosomes. Aneuploidy is often harmful and in mammals regularly leads to spontaneous abortions (miscarriages). Some aneuploid individuals are viable, for example trisomy 21 in humans, which leads to Down syndrome. Aneuploidy often alters gene dosage in ways that are detrimental to the organism; therefore, it is unlikely to spread through populations.
Polyploidy
[edit]Polyploidy, or whole genome duplication is a product of nondisjunction during meiosis which results in additional copies of the entire genome. Polyploidy is common in plants, but it has also occurred in animals, with two rounds of whole genome duplication (2R event) in the vertebrate lineage leading to humans.[4] It has also occurred in the hemiascomycete yeasts ~100 mya.[5][6]
After a whole genome duplication, there is a relatively short period of genome instability, extensive gene loss, elevated levels of nucleotide substitution and regulatory network rewiring.[7][8] In addition, gene dosage effects play a significant role.[9] Thus, most duplicates are lost within a short period, however, a considerable fraction of duplicates survive.[10] Interestingly, genes involved in regulation are preferentially retained.[11][12] Furthermore, retention of regulatory genes, most notably the Hox genes, has led to adaptive innovation.
Rapid evolution and functional divergence have been observed at the level of the transcription of duplicated genes, usually by point mutations in short transcription factor binding motifs.[13][14] Furthermore, rapid evolution of protein phosphorylation motifs, usually embedded within rapidly evolving intrinsically disordered regions is another contributing factor for survival and rapid adaptation/neofunctionalization of duplicate genes.[15] Thus, a link seems to exist between gene regulation (at least at the post-translational level) and genome evolution.[15]
Polyploidy is also a well known source of speciation, as offspring, which have different numbers of chromosomes compared to parent species, are often unable to interbreed with non-polyploid organisms. Whole genome duplications are thought to be less detrimental than aneuploidy as the relative dosage of individual genes should be the same.
As an evolutionary event
[edit]
Rate of gene duplication
[edit]Comparisons of genomes demonstrate that gene duplications are common in most species investigated. This is indicated by variable copy numbers (copy number variation) in the genome of humans[16][17] or fruit flies.[18] However, it has been difficult to measure the rate at which such duplications occur. Recent studies yielded a first direct estimate of the genome-wide rate of gene duplication in C. elegans, the first multicellular eukaryote for which such as estimate became available. The gene duplication rate in C. elegans is on the order of 10−7 duplications/gene/generation, that is, in a population of 10 million worms, one will have a gene duplication per generation. This rate is two orders of magnitude greater than the spontaneous rate of point mutation per nucleotide site in this species.[19] Older (indirect) studies reported locus-specific duplication rates in bacteria, Drosophila, and humans ranging from 10−3 to 10−7/gene/generation.[20][21][22]
Neofunctionalization
[edit]Gene duplications are an essential source of genetic novelty that can lead to evolutionary innovation. Duplication creates genetic redundancy, where the second copy of the gene is often free from selective pressure—that is, mutations of it have no deleterious effects to its host organism. If one copy of a gene experiences a mutation that affects its original function, the second copy can serve as a 'spare part' and continue to function correctly. Thus, duplicate genes accumulate mutations faster than a functional single-copy gene, over generations of organisms, and it is possible for one of the two copies to develop a new and different function. Some examples of such neofunctionalization is the apparent mutation of a duplicated digestive gene in a family of ice fish into an antifreeze gene and duplication leading to a novel snake venom gene[23] and the synthesis of 1 beta-hydroxytestosterone in pigs.[24]
Gene duplication is believed to play a major role in evolution; this stance has been held by members of the scientific community for over 100 years.[25] Susumu Ohno was one of the most famous developers of this theory in his classic book Evolution by gene duplication (1970).[26] Ohno argued that gene duplication is the most important evolutionary force since the emergence of the universal common ancestor.[27] Major genome duplication events can be quite common. It is believed that the entire yeast genome underwent duplication about 100 million years ago.[28] Plants are the most prolific genome duplicators. For example, wheat is hexaploid (a kind of polyploid), meaning that it has six copies of its genome.
Subfunctionalization
[edit]Another possible fate for duplicate genes is that both copies are equally free to accumulate degenerative mutations, so long as any defects are complemented by the other copy. This leads to a neutral "subfunctionalization" (a process of constructive neutral evolution) or DDC (duplication-degeneration-complementation) model,[29][30] in which the functionality of the original gene is distributed among the two copies. Neither gene can be lost, as both now perform important non-redundant functions, but ultimately neither is able to achieve novel functionality.
Subfunctionalization can occur through neutral processes in which mutations accumulate with no detrimental or beneficial effects. However, in some cases subfunctionalization can occur with clear adaptive benefits. If an ancestral gene is pleiotropic and performs two functions, often neither one of these two functions can be changed without affecting the other function. In this way, partitioning the ancestral functions into two separate genes can allow for adaptive specialization of subfunctions, thereby providing an adaptive benefit.[31]
Loss
[edit]Often the resulting genomic variation leads to gene dosage dependent neurological disorders such as Rett-like syndrome and Pelizaeus–Merzbacher disease.[32] Such detrimental mutations are likely to be lost from the population and will not be preserved or develop novel functions. However, many duplications are, in fact, not detrimental or beneficial, and these neutral sequences may be lost or may spread through the population through random fluctuations via genetic drift.
Identifying duplications in sequenced genomes
[edit]Criteria and single genome scans
[edit]The two genes that exist after a gene duplication event are called paralogs and usually code for proteins with a similar function and/or structure. By contrast, orthologous genes present in different species which are each originally derived from the same ancestral sequence. (See Homology of sequences in genetics).
It is important (but often difficult) to differentiate between paralogs and orthologs in biological research. Experiments on human gene function can often be carried out on other species if a homolog to a human gene can be found in the genome of that species, but only if the homolog is orthologous. If they are paralogs and resulted from a gene duplication event, their functions are likely to be too different. One or more copies of duplicated genes that constitute a gene family may be affected by insertion of transposable elements that causes significant variation between them in their sequence and finally may become responsible for divergent evolution. This may also render the chances and the rate of gene conversion between the homologs of gene duplicates due to less or no similarity in their sequences.
Paralogs can be identified in single genomes through a sequence comparison of all annotated gene models to one another. Such a comparison can be performed on translated amino acid sequences (e.g. BLASTp, tBLASTx) to identify ancient duplications or on DNA nucleotide sequences (e.g. BLASTn, megablast) to identify more recent duplications. Most studies to identify gene duplications require reciprocal-best-hits or fuzzy reciprocal-best-hits, where each paralog must be the other's single best match in a sequence comparison.[33]
Most gene duplications exist as low copy repeats (LCRs), rather highly repetitive sequences like transposable elements. They are mostly found in pericentronomic, subtelomeric and interstitial regions of a chromosome. Many LCRs, due to their size (>1Kb), similarity, and orientation, are highly susceptible to duplications and deletions.
Genomic microarrays detect duplications
[edit]Technologies such as genomic microarrays, also called array comparative genomic hybridization (array CGH), are used to detect chromosomal abnormalities, such as microduplications, in a high throughput fashion from genomic DNA samples. In particular, DNA microarray technology can simultaneously monitor the expression levels of thousands of genes across many treatments or experimental conditions, greatly facilitating the evolutionary studies of gene regulation after gene duplication or speciation.[34][35]
Next generation sequencing
[edit]Gene duplications can also be identified through the use of next-generation sequencing platforms. The simplest means to identify duplications in genomic resequencing data is through the use of paired-end sequencing reads. Tandem duplications are indicated by sequencing read pairs which map in abnormal orientations. Through a combination of increased sequence coverage and abnormal mapping orientation, it is possible to identify duplications in genomic sequencing data.
Nomenclature
[edit]
The International System for Human Cytogenomic Nomenclature (ISCN) is an international standard for human chromosome nomenclature, which includes band names, symbols and abbreviated terms used in the description of human chromosome and chromosome abnormalities. Abbreviations include dup for duplications of parts of a chromosome.[36] For example, dup(17p12) causes Charcot–Marie–Tooth disease type 1A.[37]
As amplification
[edit]Gene duplication does not necessarily constitute a lasting change in a species' genome. In fact, such changes often don't last past the initial host organism. From the perspective of molecular genetics, gene amplification is one of many ways in which a gene can be overexpressed. Genetic amplification can occur artificially, as with the use of the polymerase chain reaction technique to amplify short strands of DNA in vitro using enzymes, or it can occur naturally, as described above. If it's a natural duplication, it can still take place in a somatic cell, rather than a germline cell (which would be necessary for a lasting evolutionary change).
Role in cancer
[edit]Duplications of oncogenes are a common cause of many types of cancer. In such cases the genetic duplication occurs in a somatic cell and affects only the genome of the cancer cells themselves, not the entire organism, much less any subsequent offspring. Recent comprehensive patient-level classification and quantification of driver events in TCGA cohorts revealed that there are on average 12 driver events per tumor, of which 1.5 are amplifications of oncogenes.[38]
| Cancer type | Associated gene amplifications |
Prevalence of amplification in cancer type (percent) |
|---|---|---|
| Breast cancer | MYC | 20%[39] |
| ERBB2 (HER2) | 20%[39] | |
| CCND1 (Cyclin D1) | 15–20%[39] | |
| FGFR1 | 12%[39] | |
| FGFR2 | 12%[39] | |
| Cervical cancer | MYC | 25–50%[39] |
| ERBB2 | 20%[39] | |
| Colorectal cancer | HRAS | 30%[39] |
| KRAS | 20%[39] | |
| MYB | 15–20%[39] | |
| Esophageal cancer | MYC | 40%[39] |
| CCND1 | 25%[39] | |
| MDM2 | 13%[39] | |
| Gastric cancer | CCNE (Cyclin E) | 15%[39] |
| KRAS | 10%[39] | |
| MET | 10%[39] | |
| Glioblastoma | ERBB1 (EGFR) | 33–50%[39] |
| CDK4 | 15%[39] | |
| Head and neck cancer | CCND1 | 50%[39] |
| ERBB1 | 10%[39] | |
| MYC | 7–10%[39] | |
| Hepatocellular cancer | CCND1 | 13%[39] |
| Neuroblastoma | MYCN | 20–25%[39] |
| Ovarian cancer | MYC | 20–30%[39] |
| ERBB2 | 15–30%[39] | |
| AKT2 | 12%[39] | |
| Sarcoma | MDM2 | 10–30%[39] |
| CDK4 | 10%[39] | |
| Small cell lung cancer | MYC | 15–20%[39] |
Whole-genome duplications are also frequent in cancers, detected in 30% to 36% of tumors from the most common cancer types.[40][41] Their exact role in carcinogenesis is unclear, but they in some cases lead to loss of chromatin segregation leading to chromatin conformation changes that in turn lead to oncogenic epigenetic and transcriptional modifications.[42]
See also
[edit]References
[edit]- ^ Zhang J (2003). "Evolution by gene duplication: an update" (PDF). Trends in Ecology & Evolution. 18 (6): 292–8. doi:10.1016/S0169-5347(03)00033-8.
- ^ "Definition of Gene duplication". medterms medical dictionary. MedicineNet. 2012-03-19. Archived from the original on 2014-03-06. Retrieved 2008-12-01.
- ^ Miller, Duncan; Chen, Jianhai; Liang, Jiangtao; Betrán, Esther; Long, Manyuan; Sharakhov, Igor V. (2022-05-28). "Retrogene Duplication and Expression Patterns Shaped by the Evolution of Sex Chromosomes in Malaria Mosquitoes". Genes. 13 (6): 968. doi:10.3390/genes13060968. ISSN 2073-4425. PMC 9222922. PMID 35741730.
- ^ Dehal P, Boore JL (October 2005). "Two rounds of whole genome duplication in the ancestral vertebrate". PLOS Biology. 3 (10): e314. doi:10.1371/journal.pbio.0030314. PMC 1197285. PMID 16128622.
- ^ Wolfe, K. H.; Shields, D. C. (1997-06-12). "Molecular evidence for an ancient duplication of the entire yeast genome". Nature. 387 (6634): 708–713. Bibcode:1997Natur.387..708W. doi:10.1038/42711. ISSN 0028-0836. PMID 9192896. S2CID 4307263.
- ^ Kellis, Manolis; Birren, Bruce W.; Lander, Eric S. (2004-04-08). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature. 428 (6983): 617–624. Bibcode:2004Natur.428..617K. doi:10.1038/nature02424. ISSN 1476-4687. PMID 15004568. S2CID 4422074.
- ^ Otto, Sarah P. (2007-11-02). "The evolutionary consequences of polyploidy". Cell. 131 (3): 452–462. doi:10.1016/j.cell.2007.10.022. ISSN 0092-8674. PMID 17981114. S2CID 10054182.
- ^ Conant, Gavin C.; Wolfe, Kenneth H. (April 2006). "Functional partitioning of yeast co-expression networks after genome duplication". PLOS Biology. 4 (4): e109. doi:10.1371/journal.pbio.0040109. ISSN 1545-7885. PMC 1420641. PMID 16555924.
- ^ Papp, Balázs; Pál, Csaba; Hurst, Laurence D. (2003-07-10). "Dosage sensitivity and the evolution of gene families in yeast". Nature. 424 (6945): 194–197. Bibcode:2003Natur.424..194P. doi:10.1038/nature01771. ISSN 1476-4687. PMID 12853957. S2CID 4382441.
- ^ Lynch, M.; Conery, J. S. (2000-11-10). "The evolutionary fate and consequences of duplicate genes". Science. 290 (5494): 1151–1155. Bibcode:2000Sci...290.1151L. doi:10.1126/science.290.5494.1151. ISSN 0036-8075. PMID 11073452.
- ^ Freeling, Michael; Thomas, Brian C. (July 2006). "Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity". Genome Research. 16 (7): 805–814. doi:10.1101/gr.3681406. ISSN 1088-9051. PMID 16818725.
- ^ Davis, Jerel C.; Petrov, Dmitri A. (October 2005). "Do disparate mechanisms of duplication add similar genes to the genome?". Trends in Genetics. 21 (10): 548–551. doi:10.1016/j.tig.2005.07.008. ISSN 0168-9525. PMID 16098632.
- ^ Casneuf, Tineke; De Bodt, Stefanie; Raes, Jeroen; Maere, Steven; Van de Peer, Yves (2006). "Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana". Genome Biology. 7 (2): R13. doi:10.1186/gb-2006-7-2-r13. ISSN 1474-760X. PMC 1431724. PMID 16507168.
- ^ Li, Wen-Hsiung; Yang, Jing; Gu, Xun (November 2005). "Expression divergence between duplicate genes". Trends in Genetics. 21 (11): 602–607. doi:10.1016/j.tig.2005.08.006. ISSN 0168-9525. PMID 16140417.
- ^ a b Amoutzias, Grigoris D.; He, Ying; Gordon, Jonathan; Mossialos, Dimitris; Oliver, Stephen G.; Van de Peer, Yves (2010-02-16). "Posttranslational regulation impacts the fate of duplicated genes". Proceedings of the National Academy of Sciences of the United States of America. 107 (7): 2967–2971. Bibcode:2010PNAS..107.2967A. doi:10.1073/pnas.0911603107. ISSN 1091-6490. PMC 2840353. PMID 20080574.
- ^ Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. (July 2004). "Large-scale copy number polymorphism in the human genome". Science. 305 (5683): 525–8. Bibcode:2004Sci...305..525S. doi:10.1126/science.1098918. PMID 15273396. S2CID 20357402.
- ^ Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. (September 2004). "Detection of large-scale variation in the human genome". Nature Genetics. 36 (9): 949–51. doi:10.1038/ng1416. PMID 15286789.
- ^ Emerson JJ, Cardoso-Moreira M, Borevitz JO, Long M (June 2008). "Natural selection shapes genome-wide patterns of copy-number polymorphism in Drosophila melanogaster". Science. 320 (5883): 1629–31. Bibcode:2008Sci...320.1629E. doi:10.1126/science.1158078. PMID 18535209. S2CID 206512885.
- ^ Lipinski KJ, Farslow JC, Fitzpatrick KA, Lynch M, Katju V, Bergthorsson U (February 2011). "High spontaneous rate of gene duplication in Caenorhabditis elegans". Current Biology. 21 (4): 306–10. Bibcode:2011CBio...21..306L. doi:10.1016/j.cub.2011.01.026. PMC 3056611. PMID 21295484.
- ^ Anderson P, Roth J (May 1981). "Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons". Proceedings of the National Academy of Sciences of the United States of America. 78 (5): 3113–7. Bibcode:1981PNAS...78.3113A. doi:10.1073/pnas.78.5.3113. PMC 319510. PMID 6789329.
- ^ Watanabe Y, Takahashi A, Itoh M, Takano-Shimizu T (March 2009). "Molecular spectrum of spontaneous de novo mutations in male and female germline cells of Drosophila melanogaster". Genetics. 181 (3): 1035–43. doi:10.1534/genetics.108.093385. PMC 2651040. PMID 19114461.
- ^ Turner DJ, Miretti M, Rajan D, Fiegler H, Carter NP, Blayney ML, et al. (January 2008). "Germline rates of de novo meiotic deletions and duplications causing several genomic disorders". Nature Genetics. 40 (1): 90–5. doi:10.1038/ng.2007.40. PMC 2669897. PMID 18059269.
- ^ Lynch VJ (January 2007). "Inventing an arsenal: adaptive evolution and neofunctionalization of snake venom phospholipase A2 genes". BMC Evolutionary Biology. 7: 2. doi:10.1186/1471-2148-7-2. PMC 1783844. PMID 17233905.
- ^ Conant GC, Wolfe KH (December 2008). "Turning a hobby into a job: how duplicated genes find new functions". Nature Reviews. Genetics. 9 (12): 938–50. doi:10.1038/nrg2482. PMID 19015656. S2CID 1240225.
- ^ Taylor JS, Raes J (2004). "Duplication and divergence: the evolution of new genes and old ideas". Annual Review of Genetics. 38: 615–43. doi:10.1146/annurev.genet.38.072902.092831. PMID 15568988.
- ^ Ohno, S. (1970). Evolution by gene duplication. Springer-Verlag. ISBN 978-0-04-575015-3.
- ^ Ohno, S. (1967). Sex Chromosomes and Sex-linked Genes. Springer-Verlag. ISBN 978-91-554-5776-1.
- ^ Kellis M, Birren BW, Lander ES (April 2004). "Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae". Nature. 428 (6983): 617–24. Bibcode:2004Natur.428..617K. doi:10.1038/nature02424. PMID 15004568. S2CID 4422074.
- ^ Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (April 1999). "Preservation of duplicate genes by complementary, degenerative mutations". Genetics. 151 (4): 1531–45. doi:10.1093/genetics/151.4.1531. PMC 1460548. PMID 10101175.
- ^ Stoltzfus A (August 1999). "On the possibility of constructive neutral evolution". Journal of Molecular Evolution. 49 (2): 169–81. Bibcode:1999JMolE..49..169S. CiteSeerX 10.1.1.466.5042. doi:10.1007/PL00006540. PMID 10441669. S2CID 1743092.
- ^ Des Marais DL, Rausher MD (August 2008). "Escape from adaptive conflict after duplication in an anthocyanin pathway gene". Nature. 454 (7205): 762–5. Bibcode:2008Natur.454..762D. doi:10.1038/nature07092. PMID 18594508. S2CID 418964.
- ^ Lee JA, Lupski JR (October 2006). "Genomic rearrangements and gene copy-number alterations as a cause of nervous system disorders". Neuron. 52 (1): 103–21. doi:10.1016/j.neuron.2006.09.027. PMID 17015230. S2CID 22412305.
- ^ Hahn MW, Han MV, Han SG (November 2007). "Gene family evolution across 12 Drosophila genomes". PLOS Genetics. 3 (11): e197. doi:10.1371/journal.pgen.0030197. PMC 2065885. PMID 17997610.
- ^ Mao R, Pevsner J (2005). "The use of genomic microarrays to study chromosomal abnormalities in mental retardation". Mental Retardation and Developmental Disabilities Research Reviews. 11 (4): 279–85. doi:10.1002/mrdd.20082. PMID 16240409.
- ^ Gu X, Zhang Z, Huang W (January 2005). "Rapid evolution of expression and regulatory divergences after yeast gene duplication". Proceedings of the National Academy of Sciences of the United States of America. 102 (3): 707–12. Bibcode:2005PNAS..102..707G. doi:10.1073/pnas.0409186102. PMC 545572. PMID 15647348.
- ^ "ISCN Symbols and Abbreviated Terms". Coriell Institute for Medical Research. Retrieved 2022-10-27.
- ^ Cassandra L. Kniffin. "HARCOT-MARIE-TOOTH DISEASE, DEMYELINATING, TYPE 1A; CMT1A". OMIM. Updated : 4/23/2014
- ^ Vyatkin, Alexey D.; Otnyukov, Danila V.; Leonov, Sergey V.; Belikov, Aleksey V. (14 January 2022). "Comprehensive patient-level classification and quantification of driver events in TCGA PanCanAtlas cohorts". PLOS Genetics. 18 (1): e1009996. doi:10.1371/journal.pgen.1009996. PMC 8759692. PMID 35030162.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac Kinzler KW, Vogelstein B (2002). The genetic basis of human cancer. McGraw-Hill. p. 116. ISBN 978-0-07-137050-9.
- ^ Bielski, Craig M.; Zehir, Ahmet; Penson, Alexander V.; Donoghue, Mark T. A.; Chatila, Walid; Armenia, Joshua; Chang, Matthew T.; Schram, Alison M.; Jonsson, Philip; Bandlamudi, Chaitanya; Razavi, Pedram; Iyer, Gopa; Robson, Mark E.; Stadler, Zsofia K.; Schultz, Nikolaus (2018). "Genome doubling shapes the evolution and prognosis of advanced cancers". Nature Genetics. 50 (8): 1189–1195. doi:10.1038/s41588-018-0165-1. ISSN 1546-1718. PMC 6072608. PMID 30013179.
- ^ Quinton, Ryan J.; DiDomizio, Amanda; Vittoria, Marc A.; Kotýnková, Kristýna; Ticas, Carlos J.; Patel, Sheena; Koga, Yusuke; Vakhshoorzadeh, Jasmine; Hermance, Nicole; Kuroda, Taruho S.; Parulekar, Neha; Taylor, Alison M.; Manning, Amity L.; Campbell, Joshua D.; Ganem, Neil J. (2021). "Whole-genome doubling confers unique genetic vulnerabilities on tumour cells". Nature. 590 (7846): 492–497. Bibcode:2021Natur.590..492Q. doi:10.1038/s41586-020-03133-3. ISSN 1476-4687. PMC 7889737. PMID 33505027.
- ^ Lambuta, Ruxandra A.; Nanni, Luca; Liu, Yuanlong; Diaz-Miyar, Juan; Iyer, Arvind; Tavernari, Daniele; Katanayeva, Natalya; Ciriello, Giovanni; Oricchio, Elisa (2023-03-15). "Whole-genome doubling drives oncogenic loss of chromatin segregation". Nature. 615 (7954): 925–933. Bibcode:2023Natur.615..925L. doi:10.1038/s41586-023-05794-2. ISSN 1476-4687. PMC 10060163. PMID 36922594.
