Cell mechanics
Cell mechanics is a sub-field of biophysics that focuses on the mechanical properties and behavior of living cells and how it relates to cell function.[1] It encompasses aspects of cell biophysics, biomechanics, soft matter physics and rheology, mechanobiology and cell biology.
Eukaryotic
[edit]Eukaryotic cells [2] are cells that consist of membrane-bound organelles, a membrane-bound nucleus, and more than one linear chromosome. Being much more complex than prokaryotic cells, cells without a true nucleus, eukaryotes must protect its organelles from outside forces.
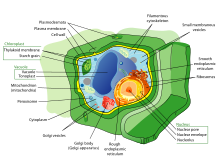
Plant
[edit]Plant cell mechanics combines principles of biomechanics and mechanobiology to investigate the growth and shaping of the plant cells. Plant cells, similar to animal cells, respond to externally applied forces, such as by reorganization of their cytoskeletal network. The presence of a considerably rigid extracellular matrix, the cell wall, however, bestows the plant cells with a set of particular properties. Mainly, the growth of plant cells is controlled by the mechanics and chemical composition of the cell wall.[3] A major part of research in plant cell mechanics is put toward the measurement and modeling of the cell wall mechanics to understand how modification of its composition and mechanical properties affects the cell function, growth and morphogenesis.[4][5]
Animal
[edit]
Because animal cells[6] do not have cell walls to protect them like plant cells, they require other specialized structures to sustain external mechanical forces. All animal cells are encased within a cell membrane made of a thin lipid bilayer that protects the cell from exposure to the outside environment. Using receptors composed of protein structures, the cell membrane is able to let selected molecules within the cell. Inside the cell membrane includes the cytoplasm, which contains the cytoskeleton.[7] A network of filamentous proteins including microtubules, intermediate filaments, and actin filaments makes up the cytoskeleton and helps maintain the cell's shape. By working together, the three types of polymers can organize themselves to counter the applied external forces and resist deformation. However, there are differences between the three polymers.
The primary structural component of the cytoskeleton is actin filaments. Being the narrowest with a diameter of 7 nm and most flexible out of the three types of polymers, actin filaments are typically found at the very edge of the cytoplasm in animal cells.[1] Formed by the linking of polymers of a protein called actin, they help give cells shape and structure and are able to transport protein packages and organelles. Furthermore, actin filaments have the ability to be assembled and disassembled quickly, allowing them to take part in cell mobility.[8]
On the other hand, intermediate filaments are more permanent structures with a diameter of 8 to 10 nm.[9] Composed of numerous fibrous protein strands wound together, intermediate proteins’ main role is bearing tension and retaining the shape and structure of the cell by securing the nucleus and other organelles in their designated areas.
The largest cytoskeletal structure of the three types of polymers is the microtubules with a diameter of 25 nm.[8] Unlike actin filaments, microtubules are stiff, hollow structures that radiate outwards from the microtubule organizing center (MTOC). Composed of tubulin proteins, microtubules are dynamic structures that allows them to shrink or grow with the addition or removal of tubulin proteins. In terms of cell mechanics, microtubules’ main purpose is to resist compressive cellular forces and act as a transportation system for motor proteins.[8]
It was shown that melanin also can have an impact on mechanic properties of cells. The research done by Sarna's team proved that heavily pigmented melanoma cells have Young's modulus about 4.93, when in non-pigmented ones it was only 0.98.[10] In another experiment they found that elasticity of melanoma cells is important for its metastasis and growth: non-pigmented tumors were bigger than pigmented and it was much easier for them to spread. They shown that there are both pigmented and non-pigmented cells in melanoma tumors, so that they can both be drug-resistant and metastatic.[10]
Measuring
[edit]Because cells are tiny, soft objects that must be measured differently than materials like metal, plastic, and glass, new techniques have been developed for the accurate measurement of cell mechanics. The variety of techniques can be divided into two categories: force application techniques and force sensing techniques.[1] In case of walled cells, such as plant or fungal cells, due to existence of a stiff, anisotropic and curved cell wall encapsulating the cells, special considerations and tailored approaches may be required compared to the methods used to measure the mechanics of animal cells.[11]
Force application
[edit]Force application techniques uses the cell's response of deformation to force applied onto the cell as a way to measure cell mechanical properties.[12] There are several different types of force application techniques including:
- Micropipette aspiration uses applied suction pressure with a small diameter glass pipet. The measurement of the length of aspiration caused by the suction pressure can reveal several cell mechanical properties.[13]
- Cantilever manipulation operates through an magnetic, electrical, or mechanical interaction between a probe and the surface of the cell that gives off a signal that can be used to measure mechanical properties.[14]
- Optical techniques involves the usage of trapped photons to manipulate cells. The photons will change in direction based on the cell's refractive index, which will cause a change in momentum, leading to a force applied upon the cell.[12]
- Mechanical techniques utilizes the incorporation of ferromagnetic beads into the cell or attached to specific receptors on the cell. When a magnetic force is applied, the stretch of the membrane can be measured to calculate mechanical properties.[12]
- Substrate strain measures elasticity through stretching the cell. The elasticity of the cell provides information that can determine motility and adhesion.[12][15]
- Compression requires the usage of applied pressure onto the entire cell. By calculating the changes of the cell's shape, compression is a way to measure mechanical responses to force.[12]
- Flow technique uses Reynold's number, a dimensionless number in fluid mechanics, to distinguish whether the cell is subject to laminar, transitional, or turbulent flow.[12]
- Acoustic force spectroscopy can be used to extract mechanical properties of single cells.[16]
Force sensing
[edit]- Wrinkling membranes requires putting the cell into a flexible silicon envelope. As the cell contracts, the magnitude of the forces can be estimated by utilizing the length and number of wrinkles.[12]
- Traction force microscopy[17] detects deformations through comparison of images the movement of fluorescent beads that have been adhered to the cell.[18] A user-friendly software package is available for download and is free of charge. It includes both fast Fourier transform traction cytometry and parameter-free Bayesian Fourier transform traction cytometry.[19]
- Cantilever sensing can detect surface stresses with the attachment of micromechanical beams on one end of the cell.[20]
- Bioreactors allow the measurement of multicellular forces in a three-dimensional system, while external forces are applied at the same time. This enables better results and more accurate data from complex experiments.[12]
- When adherent cells are excited by acoustic waves, they start to generate acoustic microstreaming flow. The velocity magnitude of this flow near the cell membrane is directly proportional to the stiffness (i.e., modulus of elasticity) of the cell.[21]
Research
[edit]Researchers who study cell mechanics are interested in the mechanics and dynamics of the assemblies and structures that make up the cell including membranes, cytoskeleton, organelles, and cytoplasm, and how they interact to give rise to the emergent properties of the cell as a whole.[22]
A particular focus of many cell mechanical studies has been the cytoskeleton, which (in animal cells) can be thought to consist of:
- actomyosin assemblies (F-actin, myosin motors, and associated binding, nucleating, capping, stabilizing, and crosslinking proteins),
- microtubules and their associated motor proteins (kinesins and dyneins),
- intermediate filaments,
- other assemblies such as spectrins and septins.
The active non-equilibrium and non-linear rheological properties of cellular assemblies have been keen point of research in recent times.[23][24] Another point of interest has been how cell cycle-related changes in cytoskeletal activity affect global cell properties, such as intracellular pressure increase during mitotic cell rounding.[25]
References
[edit]- ^ a b c Moeendarbary E, Harris AR (2014). "Cell mechanics: principles, practices, and prospects". Wiley Interdisciplinary Reviews. Systems Biology and Medicine. 6 (5): 371–88. doi:10.1002/wsbm.1275. PMC 4309479. PMID 25269160.
- ^ "Intro to eukaryotic cells". Khan Academy.
- ^ Bidhendi AJ, Altartouri B, Gosselin FP, Geitmann A (July 2019). "Mechanical Stress Initiates and Sustains the Morphogenesis of Wavy Leaf Epidermal Cells". Cell Reports. 28 (5): 1237–1250.e6. doi:10.1016/j.celrep.2019.07.006. PMID 31365867.
- ^ Bidhendi AJ, Geitmann A (January 2016). "Relating the mechanics of the primary plant cell wall to morphogenesis". Journal of Experimental Botany. 67 (2): 449–61. doi:10.1093/jxb/erv535. PMID 26689854.
- ^ Bidhendi AJ, Geitmann A (January 2018). "Finite Element Modeling of Shape Changes in Plant Cells". Plant Physiology. 176 (1): 41–56. doi:10.1104/pp.17.01684. PMC 5761827. PMID 29229695.
- ^ McGregor J (6 August 2018). "The Parts Of An Animal Cell". Science Trends. doi:10.31988/SciTrends.24128.
- ^ Clark AG, Wartlick O, Salbreux G, Paluch EK (May 2014). "Stresses at the cell surface during animal cell morphogenesis". Current Biology. 24 (10): R484-94. doi:10.1016/j.cub.2014.03.059. PMID 24845681.
- ^ a b c "Microtubules and Filaments". Scitable by Nature Education.
- ^ "What are intermediate filaments? | MBInfo". www.mechanobio.info.
- ^ a b Sarna M, Krzykawska-Serda M, Jakubowska M, Zadlo A, Urbanska K (June 2019). "Melanin presence inhibits melanoma cell spread in mice in a unique mechanical fashion". Scientific Reports. 9 (1): 9280. Bibcode:2019NatSR...9.9280S. doi:10.1038/s41598-019-45643-9. PMC 6594928. PMID 31243305.
- ^ Bidhendi AJ, Geitmann A (July 2019). "Methods to quantify primary plant cell wall mechanics". Journal of Experimental Botany. 70 (14): 3615–3648. doi:10.1093/jxb/erz281. PMID 31301141.
- ^ a b c d e f g h Rodriguez ML, McGarry PJ, Sniadecki NJ (15 October 2013). "Review on Cell Mechanics: Experimental and Modeling Approaches". Applied Mechanics Reviews. 65 (6): 060801–060801–41. Bibcode:2013ApMRv..65f0801R. doi:10.1115/1.4025355.
- ^ "Lecture 17: Cell Mechanics" (PDF).
- ^ Jalili N (10 November 2012). "Nanomechanical Cantilever-Based Manipulation for Sensing and Imaging". Nanorobotics. Springer New York. pp. 29–40. doi:10.1007/978-1-4614-2119-1_2. ISBN 978-1-4614-2118-4.
- ^ Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, et al. (April 2012). "Cells test substrate rigidity by local contractions on submicrometer pillars". Proceedings of the National Academy of Sciences of the United States of America. 109 (14): 5328–33. Bibcode:2012PNAS..109.5328G. doi:10.1073/pnas.1119886109. PMC 3325713. PMID 22431603.
- ^ Evers, Tom M.J.; Sheikhhassani, Vahid; Haks, Mariëlle C.; Storm, Cornelis; Ottenhoff, Tom H.M.; Mashaghi, Alireza (2022). "Single-cell analysis reveals chemokine-mediated differential regulation of monocyte mechanics". iScience. 25 (1): 103555. Bibcode:2022iSci...25j3555E. doi:10.1016/j.isci.2021.103555. PMC 8693412. PMID 34988399.
- ^ Lekka, Małgorzata; Gnanachandran, Kajangi; Kubiak, Andrzej; Zieliński, Tomasz; Zemła, Joanna (November 2021). "Traction force microscopy – Measuring the forces exerted by cells" (PDF). Micron. 150: 103138. doi:10.1016/j.micron.2021.103138.
- ^ Plotnikov SV, Sabass B, Schwarz US, Waterman CM (2014). "High-resolution traction force microscopy". Quantitative Imaging in Cell Biology. Methods in Cell Biology. Vol. 123. pp. 367–94. doi:10.1016/B978-0-12-420138-5.00020-3. ISBN 9780124201385. PMC 4699589. PMID 24974038.
- ^ Huang, Yunfei; Gompper, Gerhard; Sabass, Benedikt (2020). "A Bayesian traction force microscopy method with automated denoising in a user-friendly software package". Computer Physics Communications. 256: 107313. arXiv:2005.01377. doi:10.1016/j.cpc.2020.107313.
- ^ Datar R, Kim S, Jeon S, Hesketh P, Manalis S, Boisen A, Thundat T. "Cantilever Sensors: Nanomechanical Tools for Diagnostics" (PDF).
- ^ Salari A, Appak-Baskoy S, Ezzo M, Hinz B, Kolios MC, Tsai SS (March 2020). "Dancing with the Cells: Acoustic Microflows Generated by Oscillating Cells". Small. 16 (9): e1903788. doi:10.1002/smll.201903788. PMID 31829522. S2CID 209330149.
- ^ Fletcher DA, Mullins RD (January 2010). "Cell mechanics and the cytoskeleton". Nature. 463 (7280): 485–92. Bibcode:2010Natur.463..485F. doi:10.1038/nature08908. PMC 2851742. PMID 20110992.
- ^ Mizuno D, Tardin C, Schmidt CF, Mackintosh FC (January 2007). "Nonequilibrium mechanics of active cytoskeletal networks". Science. 315 (5810): 370–3. Bibcode:2007Sci...315..370M. doi:10.1126/science.1134404. PMID 17234946. S2CID 20272334.
- ^ Guo M, Ehrlicher AJ, Jensen MH, Renz M, Moore JR, Goldman RD, et al. (August 2014). "Probing the stochastic, motor-driven properties of the cytoplasm using force spectrum microscopy". Cell. 158 (4): 822–832. doi:10.1016/j.cell.2014.06.051. PMC 4183065. PMID 25126787.
- ^ Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA (January 2011). "Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding". Nature. 469 (7329): 226–30. Bibcode:2011Natur.469..226S. doi:10.1038/nature09642. PMID 21196934. S2CID 4425308.
