Eukaryotic translation
Eukaryotic translation is the biological process by which messenger RNA is translated into proteins in eukaryotes. It consists of four phases: initiation, elongation, termination, and recapping.
Initiation
[edit]
Translation initiation is the process by which the ribosome and its associated factors bind to an mRNA and are assembled at the start codon. This process is defined as either cap-dependent, in which the ribosome binds initially at the 5' cap and then travels to the stop codon, or as cap-independent, where the ribosome does not initially bind the 5' cap.
Cap-dependent initiation
[edit]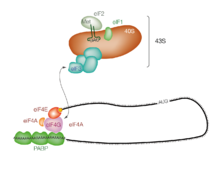
Initiation of translation usually involves the interaction of certain key proteins, the initiation factors, with a special tag bound to the 5'-end of an mRNA molecule, the 5' cap, as well as with the 5' UTR. These proteins bind the small (40S) ribosomal subunit and hold the mRNA in place.[1]
eIF3 is associated with the 40S ribosomal subunit and plays a role in keeping the large (60S) ribosomal subunit from prematurely binding. eIF3 also interacts with the eIF4F complex, which consists of three other initiation factors: eIF4A, eIF4E, and eIF4G. eIF4G is a scaffolding protein that directly associates with both eIF3 and the other two components. eIF4E is the cap-binding protein. Binding of the cap by eIF4E is often considered the rate-limiting step of cap-dependent initiation, and the concentration of eIF4E is a regulatory nexus of translational control. Certain viruses cleave a portion of eIF4G that binds eIF4E, thus preventing cap-dependent translation to hijack the host machinery in favor of the viral (cap-independent) messages. eIF4A is an ATP-dependent RNA helicase that aids the ribosome by resolving certain secondary structures formed along the mRNA transcript.[2] The poly(A)-binding protein (PABP) also associates with the eIF4F complex via eIF4G, and binds the poly-A tail of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation.[3]
This 43S preinitiation complex (43S PIC) accompanied by the protein factors moves along the mRNA chain toward its 3'-end, in a process known as 'scanning', to reach the start codon (typically AUG). In eukaryotes and archaea, the amino acid encoded by the start codon is methionine. The Met-charged initiator tRNA (Met-tRNAiMet) is brought to the P-site of the small ribosomal subunit by eukaryotic initiation factor 2 (eIF2). It hydrolyzes GTP, and signals for the dissociation of several factors from the small ribosomal subunit, eventually leading to the association of the large subunit (or the 60S subunit). The complete ribosome (80S) then commences translation elongation.
Regulation of protein synthesis is partly influenced by phosphorylation of eIF2 (via the α subunit), which is a part of the eIF2-GTP-Met-tRNAiMet ternary complex (eIF2-TC). When large numbers of eIF2 are phosphorylated, protein synthesis is inhibited. This occurs under amino acid starvation or after viral infection. However, a small fraction of this initiation factor is naturally phosphorylated. Another regulator is 4EBP, which binds to the initiation factor eIF4E and inhibits its interactions with eIF4G, thus preventing cap-dependent initiation. To oppose the effects of 4EBP, growth factors phosphorylate 4EBP, reducing its affinity for eIF4E and permitting protein synthesis.[citation needed]
While protein synthesis is globally regulated by modulating the expression of key initiation factors as well as the number of ribosomes, individual mRNAs can have different translation rates due to the presence of regulatory sequence elements. This has been shown to be important in a variety of settings including yeast meiosis and ethylene response in plants. In addition, recent work in yeast and humans suggest that evolutionary divergence in cis-regulatory sequences can impact translation regulation.[4] Additionally, RNA helicases such as DHX29 and Ded1/DDX3 may participate in the process of translation initiation, especially for mRNAs with structured 5'UTRs.[5]
Cap-independent initiation
[edit]The best-studied example of cap-independent translation initiation in eukaryotes uses the internal ribosome entry site (IRES). Unlike cap-dependent translation, cap-independent translation does not require a 5' cap to initiate scanning from the 5' end of the mRNA until the start codon. The ribosome can localize to the start site by direct binding, initiation factors, and/or ITAFs (IRES trans-acting factors) bypassing the need to scan the entire 5' UTR. This method of translation is important in conditions that require the translation of specific mRNAs during cellular stress, when overall translation is reduced. Examples include factors responding to apoptosis and stress-induced responses.[6]
Elongation
[edit]
Elongation depends on eukaryotic elongation factors. At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis. The initiator tRNA occupies the P site in the ribosome, and the A site is ready to receive an aminoacyl-tRNA. During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle. The steps in this microcycle are (1) positioning the correct aminoacyl-tRNA in the A site of the ribosome, which is brought into that site by eEF1, (2) forming the peptide bond, and (3) shifting the mRNA by one codon relative to the ribosome with the help of eEF2. Unlike bacteria, in which translation initiation occurs as soon as the 5' end of an mRNA is synthesized, in eukaryotes, such tight coupling between transcription and translation is not possible because transcription and translation are carried out in separate compartments of the cell (the nucleus and cytoplasm). Eukaryotic mRNA precursors must be processed in the nucleus (e.g., capping, polyadenylation, splicing) in ribosomes before they are exported to the cytoplasm for translation. Translation can also be affected by ribosomal pausing, which can trigger endonucleolytic attack of the tRNA, a process termed mRNA no-go decay. Ribosomal pausing also aids co-translational folding of the nascent polypeptide on the ribosome, and delays protein translation while it is encoding tRNA. This can trigger ribosomal frameshifting.[7]
Termination
[edit]Termination of elongation depends on eukaryotic release factors. The process is similar to that of bacterial termination, but unlike bacterial termination, there is a universal release factor, eRF1, that recognizes all three stop codons. Upon termination, the ribosome is disassembled and the completed polypeptide is released. eRF3 is a ribosome-dependent GTPase that helps eRF1 release the completed polypeptide. The human genome encodes a few genes whose mRNA stop codon are surprisingly leaky: In these genes, termination of translation is inefficient due to special RNA bases in the vicinity of the stop codon. Leaky termination in these genes leads to translational readthrough of up to 10% of the stop codons of these genes. Some of these genes encode functional protein domains in their readthrough extension so that new protein isoforms can arise. This process has been termed 'functional translational readthrough'.[8]
Regulation and modification of translation
[edit]Translation is one of the key energy consumers in cells, hence it is strictly regulated. Numerous mechanisms have evolved that control and regulate translation in eukaryotes as well as prokaryotes. Regulation of translation can impact the global rate of protein synthesis which is closely coupled to the metabolic and proliferative state of a cell. To delve deeper into this intricate process, scientists typically use a technique known as ribosome profiling.[9] This method enables researchers to take a snapshot of the translatome, showing which parts of the mRNA are being translated into proteins by ribosomes at a given time. Ribosome profiling provides valuable insights into translation dynamics, revealing the complex interplay between gene sequence, mRNA structure, and translation regulation. Expanding on this concept, a more recent development is single-cell ribosome profiling, a technique that allows us to study the translation process at the resolution of individual cells.[10] Single-cell ribosome profiling has the potential to shed light on the heterogeneous nature of cells, leading to a more nuanced understanding of how translation regulation can impact cell behavior, metabolic state, and responsiveness to various stimuli or conditions.
Amino acid substitution
[edit]In some cells certain amino acids can be depleted and thus affect translation efficiency. For instance, activated T cells secrete interferon-γ which triggers intracellular tryptophan shortage by upregulating the indoleamine 2,3-dioxygenase 1 (IDO1) enzyme. Surprisingly, despite tryptophan depletion, in-frame protein synthesis continues across tryptophan codons. This is achieved by incorporation of phenylalanine instead of tryptophan. The resulting peptides are called W>F "substitutants". Such W>F substitutants are abundant in certain cancer types and have been associated with increased IDO1 expression. Functionally, W>F substitutants can impair protein activity.[11]
See also
[edit]References
[edit]- ^ Malys N, McCarthy JE (March 2011). "Translation initiation: variations in the mechanism can be anticipated". Cellular and Molecular Life Sciences. 68 (6): 991–1003. doi:10.1007/s00018-010-0588-z. PMC 11115079. PMID 21076851. S2CID 31720000.
- ^ Hellen CU, Sarnow P (July 2001). "Internal ribosome entry sites in eukaryotic mRNA molecules". Genes & Development. 15 (13): 1593–612. doi:10.1101/gad.891101. PMID 11445534.
- ^ Wells SE, Hillner PE, Vale RD, Sachs AB (July 1998). "Circularization of mRNA by eukaryotic translation initiation factors". Molecular Cell. 2 (1): 135–40. doi:10.1016/S1097-2765(00)80122-7. PMID 9702200.
- ^ Cenik C, Cenik ES, Byeon GW, Grubert F, Candille SI, Spacek D, Alsallakh B, Tilgner H, Araya CL, Tang H, Ricci E, Snyder MP (November 2015). "Integrative analysis of RNA, translation, and protein levels reveals distinct regulatory variation across humans". Genome Research. 25 (11): 1610–21. doi:10.1101/gr.193342.115. PMC 4617958. PMID 26297486.
- ^ Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV (December 2008). "Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29". Cell. 135 (7): 1237–50. doi:10.1016/j.cell.2008.10.037. PMC 2948571. PMID 19109895.
- ^ López-Lastra M, Rivas A, Barría MI (2005). "Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation". Biological Research. 38 (2–3): 121–46. doi:10.4067/s0716-97602005000200003. PMID 16238092.
- ^ Buchan JR, Stansfield I (September 2007). "Halting a cellular production line: responses to ribosomal pausing during translation". Biology of the Cell. 99 (9): 475–87. doi:10.1042/BC20070037. PMID 17696878.
- ^ Schueren F, Thoms S (August 2016). "Functional Translational Readthrough: A Systems Biology Perspective". PLOS Genetics. 12 (8): e1006196. doi:10.1371/JOURNAL.PGEN.1006196. PMC 4973966. PMID 27490485.
- ^ Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS (April 2009). "Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling". Science. 324 (5924): 218–23. Bibcode:2009Sci...324..218I. doi:10.1126/science.1168978. PMC 2746483. PMID 19213877.
- ^ Ozadam H, Tonn T, Han CM, Segura A, Hoskins I, Rao S; et al. (2023). "Single-cell quantification of ribosome occupancy in early mouse development". Nature. 618 (7967): 1057–1064. Bibcode:2023Natur.618.1057O. doi:10.1038/s41586-023-06228-9. PMC 10307641. PMID 37344592.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pataskar, Abhijeet; Champagne, Julien; Nagel, Remco; Kenski, Juliana; Laos, Maarja; Michaux, Justine; Pak, Hui Song; Bleijerveld, Onno B.; Mordente, Kelly; Navarro, Jasmine Montenegro; Blommaert, Naomi (2022-03-24). "Tryptophan depletion results in tryptophan-to-phenylalanine substitutants". Nature. 603 (7902): 721–727. Bibcode:2022Natur.603..721P. doi:10.1038/s41586-022-04499-2. ISSN 0028-0836. PMC 8942854. PMID 35264796.
