Helicobacter pylori
| Helicobacter pylori | |
|---|---|
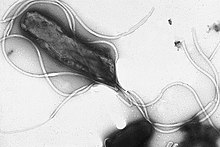
| |
| Electron micrograph of H. pylori possessing multiple flagella (negative staining) | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Campylobacterota |
| Class: | "Campylobacteria" |
| Order: | Campylobacterales |
| Family: | Helicobacteraceae |
| Genus: | Helicobacter |
| Species: | H. pylori
|
| Binomial name | |
| Helicobacter pylori (Marshall et al. 1985) Goodwin et al., 1989
| |
| Synonyms | |
| |
Helicobacter pylori, previously known as Campylobacter pylori, is a gram-negative, flagellated, helical bacterium. Mutants can have a rod or curved rod shape that exhibits less virulence.[1][2] Its helical body (from which the genus name Helicobacter derives) is thought to have evolved to penetrate the mucous lining of the stomach, helped by its flagella, and thereby establish infection.[3][2] The bacterium was first identified as the causal agent of gastric ulcers in 1983 by Australian physician-scientists Barry Marshall and Robin Warren.[4][5] In 2005, they were awarded the Nobel Prize in Physiology or Medicine for their discovery.[6]
Infection of the stomach with H. pylori is not the cause of illness itself: over half of the global population is infected, but most individuals are asymptomatic.[7][8] Persistent colonization with more virulent strains can induce a number of gastric and non-gastric disorders.[9] Gastric disorders due to infection begin with gastritis, or inflammation of the stomach lining.[10] When infection is persistent, the prolonged inflammation will become chronic gastritis. Initially, this will be non-atrophic gastritis, but the damage caused to the stomach lining can bring about the development of atrophic gastritis and ulcers within the stomach itself or the duodenum (the nearest part of the intestine).[10] At this stage, the risk of developing gastric cancer is high.[11] However, the development of a duodenal ulcer confers a comparatively lower risk of cancer.[12] Helicobacter pylori is a class 1 carcinogenic bacteria, and potential cancers include gastric MALT lymphoma and gastric cancer.[10][11] Infection with H. pylori is responsible for an estimated 89% of all gastric cancers and is linked to the development of 5.5% of all cases cancers worldwide.[13][14] H. pylori is the only bacterium known to cause cancer.[15]
Extragastric complications that have been linked to H. pylori include anemia due either to iron deficiency or vitamin B12 deficiency, diabetes mellitus, cardiovascular illness, and certain neurological disorders.[16] An inverse association has also been claimed with H. pylori having a positive protective effect against asthma, esophageal cancer, inflammatory bowel disease (including gastroesophageal reflux disease and Crohn's disease), and others.[16]
Some studies suggest that H. pylori plays an important role in the natural stomach ecology by influencing the type of bacteria that colonize the gastrointestinal tract.[17][18] Other studies suggest that non-pathogenic strains of H. pylori may beneficially normalize stomach acid secretion, and regulate appetite.[19]
In 2023, it was estimated that about two-thirds of the world's population was infected with H. pylori, being more common in developing countries.[20] The prevalence has declined in many countries due to eradication treatments with antibiotics and proton-pump inhibitors, and with increased standards of living.[21][22]
Microbiology
[edit]Helicobacter pylori is a species of gram-negative bacteria in the Helicobacter genus.[23] About half the world's population is infected with H. pylori but only a few strains are pathogenic. H pylori is a helical bacterium having a predominantly helical shape, also often described as having a spiral or S shape.[24][25] Its helical shape is better suited for progressing through the viscous mucosa lining of the stomach, and is maintained by a number of enzymes in the cell wall's peptidoglycan.[1] The bacteria reach the less acidic mucosa by use of their flagella.[26] Three strains studied showed a variation in length from 2.8–3.3 μm but a fairly constant diameter of 0.55–0.58 μm.[24] H. pylori can convert from a helical to an inactive coccoid form that can evade the immune system, and that may possibly become viable, known as viable but nonculturable (VBNC).[27][28]
Helicobacter pylori is microaerophilic – that is, it requires oxygen, but at lower concentration than in the atmosphere. It contains a hydrogenase that can produce energy by oxidizing molecular hydrogen (H2) made by intestinal bacteria.[29]
H. pylori can be demonstrated in tissue by Gram stain, Giemsa stain, H&E stain, Warthin-Starry silver stain, acridine orange stain, and phase-contrast microscopy. It is capable of forming biofilms. Biofilms help to hinder the action of antibiotics and can contribute to treatment failure.[30][31]
To successfully colonize its host, H. pylori uses many different virulence factors including oxidase, catalase, and urease.[32] Urease is the most abundant protein, its expression representing about 10% of the total protein weight.[33]
H. pylori possesses five major outer membrane protein families.[32] The largest family includes known and putative adhesins. The other four families are porins, iron transporters, flagellum-associated proteins, and proteins of unknown function. Like other typical gram-negative bacteria, the outer membrane of H. pylori consists of phospholipids and lipopolysaccharide (LPS). The O-antigen of LPS may be fucosylated and mimic Lewis blood group antigens found on the gastric epithelium.[32]
Genome
[edit]Helicobacter pylori consists of a large diversity of strains, and hundreds of genomes have been completely sequenced.[34][35][36] The genome of the strain 26695 consists of about 1.7 million base pairs, with some 1,576 genes.[37][38] The pan-genome, that is the combined set of 30 sequenced strains, encodes 2,239 protein families (orthologous groups OGs).[39] Among them, 1,248 OGs are conserved in all the 30 strains, and represent the universal core. The remaining 991 OGs correspond to the accessory genome in which 277 OGs are unique to one strain.[40]
There are eleven restriction modification systems in the genome of H. pylori.[38] This is an unusually high number providing a defence against bacteriophages.[38]
Transcriptome
[edit]Single-cell transcriptomics using single-cell RNA-Seq gave the complete transcriptome of H. pylori which was published in 2010. This analysis of its transcription confirmed the known acid induction of major virulence loci, including the urease (ure) operon and the Cag pathogenicity island (PAI).[41] A total of 1,907 transcription start sites 337 primary operons, and 126 additional suboperons, and 66 monocistrons were identified. Until 2010, only about 55 transcription start sites (TSSs) were known in this species. 27% of the primary TSSs are also antisense TSSs, indicating that – similar to E. coli – antisense transcription occurs across the entire H. pylori genome. At least one antisense TSS is associated with about 46% of all open reading frames, including many housekeeping genes.[41] About 50% of the 5′ UTRs (leader sequences) are 20–40 nucleotides (nt) in length and support the AAGGag motif located about 6 nt (median distance) upstream of start codons as the consensus Shine–Dalgarno sequence in H. pylori.[41]
Proteome
[edit]The proteome of H. pylori has been systematically analyzed and more than 70% of its proteins have been detected by mass spectrometry, and other methods. About 50% of the proteome has been quantified, informing of the number of protein copies in a typical cell.[42]
Studies of the interactome have identified more than 3000 protein-protein interactions. This has provided information of how proteins interact with each other, either in stable protein complexes or in more dynamic, transient interactions, which can help to identify the functions of the protein. This in turn helps researchers to find out what the function of uncharacterized proteins is, e.g. when an uncharacterized protein interacts with several proteins of the ribosome (that is, it is likely also involved in ribosome function). About a third of all ~1,500 proteins in H. pylori remain uncharacterized and their function is largely unknown.[43]
Infection
[edit]
An infection with Helicobacter pylori can either have no symptoms even when lasting a lifetime, or can harm the stomach and duodenal linings by inflammatory responses induced by several mechanisms associated with a number of virulence factors. Colonization can initially cause H. pylori induced gastritis, an inflammation of the stomach lining that became a listed disease in ICD11.[44][45][46] This will progress to chronic gastritis if left untreated. Chronic gastritis may lead to atrophy of the stomach lining, and the development of peptic ulcers (gastric or duodenal). These changes may be seen as stages in the development of gastric cancer, known as Correa's cascade.[47][48] Extragastric complications that have been linked to H. pylori include anemia due either to iron-deficiency or vitamin B12 deficiency, diabetes mellitus, cardiovascular, and certain neurological disorders.[16]
Peptic ulcers are a consequence of inflammation that allows stomach acid and the digestive enzyme pepsin to overwhelm the protective mechanisms of the mucous membranes. The location of colonization of H. pylori, which affects the location of the ulcer, depends on the acidity of the stomach.[49] In people producing large amounts of acid, H. pylori colonizes near the pyloric antrum (exit to the duodenum) to avoid the acid-secreting parietal cells at the fundus (near the entrance to the stomach).[32] G cells express relatively high levels of PD-L1 that protects these cells from H. pylori-induced immune destruction.[50] In people producing normal or reduced amounts of acid, H. pylori can also colonize the rest of the stomach.
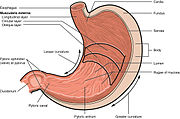
The inflammatory response caused by bacteria colonizing near the pyloric antrum induces G cells in the antrum to secrete the hormone gastrin, which travels through the bloodstream to parietal cells in the fundus.[51] Gastrin stimulates the parietal cells to secrete more acid into the stomach lumen, and over time increases the number of parietal cells, as well.[52] The increased acid load damages the duodenum, which may eventually lead to the formation of ulcers.
Helicobacter pylori is a class I carcinogen, and potential cancers include gastric mucosa-associated lymphoid tissue (MALT) lymphomas and gastric cancer.[10][11][53] Less commonly, diffuse large B-cell lymphoma of the stomach is a risk.[54] Infection with H. pylori is responsible for around 89 per cent of all gastric cancers, and is linked to the development of 5.5 per cent of all cases of cancer worldwide.[13][14] Although the data varies between different countries, overall about 1% to 3% of people infected with Helicobacter pylori develop gastric cancer in their lifetime compared to 0.13% of individuals who have had no H. pylori infection.[55][32] H. pylori-induced gastric cancer is the third highest cause of worldwide cancer mortality as of 2018.[56] Because of the usual lack of symptoms, when gastric cancer is finally diagnosed it is often fairly advanced. More than half of gastric cancer patients have lymph node metastasis when they are initially diagnosed.[57]

Chronic inflammation that is a feature of cancer development is characterized by infiltration of neutrophils and macrophages to the gastric epithelium, which favors the accumulation of pro-inflammatory cytokines, reactive oxygen species (ROS) and reactive nitrogen species (RNS) that cause DNA damage.[58] The oxidative DNA damage and levels of oxidative stress can be indicated by a biomarker, 8-oxo-dG.[58][59] Other damage to DNA includes double-strand breaks.[60]
Small gastric and colorectal polyps are adenomas that are more commonly found in association with the mucosal damage induced by H. pylori gastritis.[61][62] Larger polyps can in time become cancerous.[63][61] A modest association of H. pylori has been made with the development of colorectal cancers, but as of 2020 causality had yet to be proved.[64][63]
Signs and symptoms
[edit]Most people infected with H. pylori never experience any symptoms or complications, but will have a 10% to 20% risk of developing peptic ulcers or a 0.5% to 2% risk of stomach cancer.[8][65] H. pylori induced gastritis may present as acute gastritis with stomach ache, nausea, and ongoing dyspepsia (indigestion) that is sometimes accompanied by depression and anxiety.[8][66] Where the gastritis develops into chronic gastritis, or an ulcer, the symptoms are the same and can include indigestion, stomach or abdominal pains, nausea, bloating, belching, feeling hunger in the morning, feeling full too soon, and sometimes vomiting, heartburn, bad breath, and weight loss.[67][68]
Complications of an ulcer can cause severe signs and symptoms such as black or tarry stool indicative of bleeding into the stomach or duodenum; blood - either red or coffee-ground colored in vomit; persistent sharp or severe abdominal pain; dizziness, and a fast heartbeat.[67][68] Bleeding is the most common complication. In cases caused by H. pylori there was a greater need for hemostasis often requiring gastric resection.[69] Prolonged bleeding may cause anemia leading to weakness and fatigue. Inflammation of the pyloric antrum, which connects the stomach to the duodenum, is more likely to lead to duodenal ulcers, while inflammation of the corpus may lead to a gastric ulcer.
Stomach cancer can cause nausea, vomiting, diarrhoea, constipation, and unexplained weight loss.[70] Gastric polyps are adenomas that are usually asymptomatic and benign, but may be the cause of dyspepsia, heartburn, bleeding from the stomach, and, rarely, gastric outlet obstruction.[61][71] Larger polyps may have become cancerous.[61] Colorectal polyps may be the cause of rectal bleeding, anemia, constipation, diarrhea, weight loss, and abdominal pain.[72]
Pathophysiology
[edit]Virulence factors help a pathogen to evade the immune response of the host, and to successfully colonize. The many virulence factors of H. pylori include its flagella, the production of urease, adhesins, serine protease HtrA (high temperature requirement A), and the major exotoxins CagA and VacA.[30][73] The presence of VacA and CagA are associated with more advanced outcomes.[74] CagA is an oncoprotein associated with the development of gastric cancer.[7]

H. pylori infection is associated with epigenetically reduced efficiency of the DNA repair machinery, which favors the accumulation of mutations and genomic instability as well as gastric carcinogenesis.[75] It has been shown that expression of two DNA repair proteins, ERCC1 and PMS2, was severely reduced once H. pylori infection had progressed to cause dyspepsia.[76] Dyspepsia occurs in about 20% of infected individuals.[77] Epigenetically reduced protein expression of DNA repair proteins MLH1, MGMT and MRE11 are also evident. Reduced DNA repair in the presence of increased DNA damage increases carcinogenic mutations and is likely a significant cause of gastric carcinogenesis.[59][78][79] These epigenetic alterations are due to H. pylori-induced methylation of CpG sites in promoters of genes[78] and H. pylori-induced altered expression of multiple microRNAs.[79]
Two related mechanisms by which H. pylori could promote cancer have been proposed. One mechanism involves the enhanced production of free radicals near H. pylori and an increased rate of host cell mutation. The other proposed mechanism has been called a "perigenetic pathway",[80] and involves enhancement of the transformed host cell phenotype by means of alterations in cell proteins, such as adhesion proteins. H. pylori has been proposed to induce inflammation and locally high levels of tumor necrosis factor (TNF), also known as tumor necrosis factor alpha (TNFα)), and/or interleukin 6 (IL-6).[81] According to the proposed perigenetic mechanism, inflammation-associated signaling molecules, such as TNF, can alter gastric epithelial cell adhesion and lead to the dispersion and migration of mutated epithelial cells without the need for additional mutations in tumor suppressor genes, such as genes that code for cell adhesion proteins.[82]
Flagellum
[edit]The first virulence factor of Helicobacter pylori that enables colonization is its flagellum.[83] H. pylori has from two to seven flagella at the same polar location which gives it a high motility. The flagellar filaments are about 3 μm long, and composed of two copolymerized flagellins, FlaA and FlaB, coded by the genes flaA, and flaB.[26][73] The minor flagellin FlaB is located in the proximal region and the major flagellin FlaA makes up the rest of the flagellum.[84] The flagella are sheathed in a continuation of the bacterial outer membrane which gives protection against the gastric acidity. The sheath is also the location of the origin of the outer membrane vesicles that gives protection to the bacterium from bacteriophages.[84]
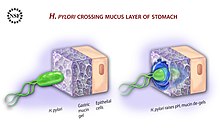
Flagella motility is provided by the proton motive force provided by urease-driven hydrolysis allowing chemotactic movements towards the less acidic pH gradient in the mucosa.[30] The mucus layer is about 300 μm thick, and the helical shape of H. pylori aided by its flagella helps it to burrow through this layer where it colonises a narrow region of about 25 μm closest to the epithelial cell layer, where the pH is near to neutral. They further colonise the gastric pits and live in the gastric glands.[1][84][85] Occasionally the bacteria are found inside the epithelial cells themselves.[86] The use of quorum sensing by the bacteria enables the formation of a biofilm which furthers persistent colonisation. In the layers of the biofilm, H. pylori can escape from the actions of antibiotics, and also be protected from host-immune responses.[87][88] In the biofilm, H. pylori can change the flagella to become adhesive structures.[89]
Urease
[edit]
In addition to using chemotaxis to avoid areas of high acidity (low pH), H. pylori also produces large amounts of urease, an enzyme which breaks down the urea present in the stomach to produce ammonia and bicarbonate, which are released into the bacterial cytosol and the surrounding environment, creating a neutral area.[90] The decreased acidity (higher pH) changes the mucus layer from a gel-like state to a more viscous state that makes it easier for the flagella to move the bacteria through the mucosa and attach to the gastric epithelial cells.[90] Helicobacter pylori is one of the few known types of bacterium that has a urea cycle which is uniquely configured in the bacterium.[91] 10% of the cell is of nitrogen, a balance that needs to be maintained. Any excess is stored in urea excreted in the urea cycle.[91]
A final stage enzyme in the urea cycle is arginase, an enzyme that is crucial to the pathogenesis of H. pylori. Arginase produces ornithine and urea, which the enzyme urease breaks down into carbonic acid and ammonia. Urease is the bacterium’s most abundant protein, accounting for 10–15% of the bacterium's total protein content. Its expression is not only required for establishing initial colonization in the breakdown of urea to carbonic acid and ammonia, but is also essential for maintaining chronic infection.[92][65] Ammonia reduces stomach acidity, allowing the bacteria to become locally established. Arginase promotes the persistence of infection by consuming arginine; arginine is used by macrophages to produce nitric oxide, which has a strong antimicrobial effect.[91][93] The ammonia produced to regulate pH is toxic to epithelial cells.[94]
Adhesins
[edit]H. pylori must make attachment with the epithelial cells to prevent its being swept away with the constant movement and renewal of the mucus. To give them this adhesion, bacterial outer membrane proteins as virulence factors called adhesins are produced.[95] BabA (blood group antigen binding adhesin) is most important during initial colonization, and SabA (sialic acid binding adhesin) is important in persistence. BabA attaches to glycans and mucins in the epithelium.[95] BabA (coded for by the babA2 gene) also binds to the Lewis b antigen displayed on the surface of the epithelial cells.[96] Adherence via BabA is acid sensitive and can be fully reversed by a decreased pH. It has been proposed that BabA's acid responsiveness enables adherence while also allowing an effective escape from an unfavorable environment such as a low pH that is harmful to the organism.[97] SabA (coded for by the sabA gene) binds to increased levels of sialyl-Lewis X antigen expressed on gastric mucosa.[98]
Cholesterol glucoside
[edit]The outer membrane contains cholesterol glucoside, a sterol glucoside that H. pylori glycosylates from the cholesterol in the gastric gland cells, and inserts it into its outer membrane.[99] This cholesterol glucoside is important for membrane stability, morphology and immune evasion, and is rarely found in other bacteria.[100][101]
The enzyme responsible for this is cholesteryl α-glucosyltransferase (αCgT or Cgt), encoded by the HP0421 gene.[102] A major effect of the depletion of host cholesterol by Cgt is to disrupt cholesterol-rich lipid rafts in the epithelial cells. Lipid rafts are involved in cell signalling and their disruption causes a reduction in the immune inflammatory response, particularly by reducing interferon gamma.[103] Cgt is also secreted by the type IV secretion system, and is secreted in a selective way so that gastric niches where the pathogen can thrive are created.[102] Its lack has been shown to give vulnerability from environmental stress to bacteria, and also to disrupt CagA-mediated interactions.[99]
Catalase
[edit]Colonization induces an intense anti-inflammatory response as a first-line immune system defence. Phagocytic leukocytes and monocytes infiltrate the site of infection, and antibodies are produced.[104] H. pylori is able to adhere to the surface of the phagocytes and impede their action. This is responded to by the phagocyte in the generation and release of oxygen metabolites into the surrounding space. H. pylori can survive this response by the activity of catalase at its attachment to the phagocytic cell surface. Catalase decomposes hydrogen peroxide into water and oxygen, protecting the bacteria from toxicity. Catalase has been shown to almost completely inhibit the phagocytic oxidative response.[104] It is coded for by the gene katA.[105]
Tipα
[edit]TNF-inducing protein alpha (Tipα) is a carcinogenic protein encoded by HP0596 unique to H. pylori that induces the expression of tumor necrosis factor.[82][106] Tipα enters gastric cancer cells where it binds to cell surface nucleolin, and induces the expression of vimentin. Vimentin is important in the epithelial–mesenchymal transition associated with the progression of tumors.[107]
CagA
[edit]CagA (cytotoxin-associated antigen A) is a major virulence factor for H. pylori, an oncoprotein that is encoded by the cagA gene. Bacterial strains with the cagA gene are associated with the ability to cause ulcers, MALT lymphomas, and gastric cancer.[108][109] The cagA gene codes for a relatively long (1186-amino acid) protein. The cag pathogenicity island (PAI) has about 30 genes, part of which code for a complex type IV secretion system (T4SS or TFSS). The low GC-content of the cag PAI relative to the rest of the Helicobacter genome suggests the island was acquired by horizontal transfer from another bacterial species.[38] The serine protease HtrA also plays a major role in the pathogenesis of H. pylori. The HtrA protein enables the bacterium to transmigrate across the host cells' epithelium, and is also needed for the translocation of CagA.[110]
The virulence of H. pylori may be increased by genes of the cag pathogenicity island; about 50–70% of H. pylori strains in Western countries carry it.[111] Western people infected with strains carrying the cag PAI have a stronger inflammatory response in the stomach and are at a greater risk of developing peptic ulcers or stomach cancer than those infected with strains lacking the island.[32] Following attachment of H. pylori to stomach epithelial cells, the type IV secretion system expressed by the cag PAI "injects" the inflammation-inducing agent, peptidoglycan, from their own cell walls into the epithelial cells. The injected peptidoglycan is recognized by the cytoplasmic pattern recognition receptor (immune sensor) Nod1, which then stimulates expression of cytokines that promote inflammation.[112]
The type-IV secretion apparatus also injects the cag PAI-encoded protein CagA into the stomach's epithelial cells, where it disrupts the cytoskeleton, adherence to adjacent cells, intracellular signaling, cell polarity, and other cellular activities.[113] Once inside the cell, the CagA protein is phosphorylated on tyrosine residues by a host cell membrane-associated tyrosine kinase (TK). CagA then allosterically activates protein tyrosine phosphatase/protooncogene Shp2.[114] These proteins are directly toxic to cells lining the stomach and signal strongly to the immune system that an invasion is under way. As a result of the bacterial presence, neutrophils and macrophages set up residence in the tissue to fight the bacteria assault.[115] Pathogenic strains of H. pylori have been shown to activate the epidermal growth factor receptor (EGFR), a membrane protein with a TK domain. Activation of the EGFR by H. pylori is associated with altered signal transduction and gene expression in host epithelial cells that may contribute to pathogenesis. A C-terminal region of the CagA protein (amino acids 873–1002) has also been suggested to be able to regulate host cell gene transcription, independent of protein tyrosine phosphorylation.[109] A great deal of diversity exists between strains of H. pylori, and the strain that infects a person can predict the outcome.
VacA
[edit]VacA (vacuolating cytotoxin autotransporter) is another major virulence factor encoded by the vacA gene.[116] All strains of H. pylori carry this gene but there is much diversity, and only 50% produce the encoded cytotoxin.[92][33] The four main subtypes of vacA are s1/m1, s1/m2, s2/m1, and s2/m2. s1/m1 and s1/m2 are known to cause an increased risk of gastric cancer.[117] VacA is an oligomeric protein complex that causes a progressive vacuolation in the epithelial cells leading to their death.[118] The vacuolation has also been associated with promoting intracellular reservoirs of H. pylori by disrupting the calcium channel cell membrane TRPML1.[119] VacA has been shown to increase the levels of COX2, an up-regulation that increases the production of a prostaglandin indicating a strong host cell inflammatory response.[118][120]
Outer membrane proteins and vesicles
[edit]About 4% of the genome encodes for outer membrane proteins that can be grouped into five families.[121] The largest family includes bacterial adhesins. The other four families are porins, iron transporters, flagellum-associated proteins, and proteins of unknown function. Like other typical gram-negative bacteria, the outer membrane of H. pylori consists of phospholipids and lipopolysaccharide (LPS). The O-antigen of LPS may be fucosylated and mimic Lewis blood group antigens found on the gastric epithelium.[32]
H. pylori forms blebs from the outer membrane that pinch off as outer membrane vesicles to provide an alternative delivery system for virulence factors including CagA.[99]
A Helicobacter cysteine-rich protein HcpA is known to trigger an immune response, causing inflammation.[122] A Helicobacter pylori virulence factor DupA is associated with the development of duodenal ulcers.[123]
Mechanisms of tolerance
[edit]The need for survival has led to the development of different mechanisms of tolerance that enable the persistence of H. pylori.[124] These mechanisms can also help to overcome the effects of antibiotics.[124] H. pylori has to not only survive the harsh gastric acidity but also the sweeping of mucus by continuous peristalsis, and phagocytic attack accompanied by the release of reactive oxygen species.[125] All organisms encode genetic programs for response to stressful conditions including those that cause DNA damage.[126] Stress conditions activate bacterial response mechanisms that are regulated by proteins expressed by regulator genes.[124] The oxidative stress can induce potentially lethal mutagenic DNA adducts in its genome. Surviving this DNA damage is supported by transformation-mediated recombinational repair, that contributes to successful colonization.[127][128] H. pylori is naturally competent for transformation. While many organisms are competent only under certain environmental conditions, such as starvation, H. pylori is competent throughout logarithmic growth.[126]
Transformation (the transfer of DNA from one bacterial cell to another through the intervening medium) appears to be part of an adaptation for DNA repair.[126] Homologous recombination is required for repairing double-strand breaks (DSBs). The AddAB helicase-nuclease complex resects DSBs and loads RecA onto single-strand DNA (ssDNA), which then mediates strand exchange, leading to homologous recombination and repair. The requirement of RecA plus AddAB for efficient gastric colonization suggests that H. pylori is either exposed to double-strand DNA damage that must be repaired or requires some other recombination-mediated event. In particular, natural transformation is increased by DNA damage in H. pylori, and a connection exists between the DNA damage response and DNA uptake in H. pylori.[126] This natural competence contributes to the persistence of H. pylori. H. pylori has much greater rates of recombination and mutation than other bacteria.[3] Genetically different strains can be found in the same host, and also in different regions of the stomach.[129] An overall response to multiple stressors can result from an interaction of the mechanisms.[124]
RuvABC proteins are essential to the process of recombinational repair, since they resolve intermediates in this process termed Holliday junctions. H. pylori mutants that are defective in RuvC have increased sensitivity to DNA-damaging agents and to oxidative stress, exhibit reduced survival within macrophages, and are unable to establish successful infection in a mouse model.[130] Similarly, RecN protein plays an important role in DSB repair.[131] An H. pylori recN mutant displays an attenuated ability to colonize mouse stomachs, highlighting the importance of recombinational DNA repair in survival of H. pylori within its host.[131]
Biofilm
[edit]An effective sustained colonization response is the formation of a biofilm. Having first adhered to cellular surfaces, the bacteria produce and secrete extracellular polymeric substance (EPS). EPS consists largely of biopolymers and provides the framework for the biofilm structure.[90] H. pylori helps the biofilm formation by altering its flagella into adhesive structures that provide adhesion between the cells.[89] Layers of aggregated bacteria as microcolonies accumulate to thicken the biofilm.
The matrix of EPS prevents the entry of antibiotics and immune cells, and provides protection from heat and competition from other microorganisms.[90] Channels form between the cells in the biofilm matrix allowing the transport of nutrients, enzymes, metabolites, and waste.[90] Cells in the deep layers may be nutritionally deprived and enter into the coccoid dormant-like state.[132][133] By changing the shape of the bacterium to a coccoid form, the exposure of LPS (targeted by antibiotics) becomes limited, and so evades detection by the immune system.[134] It has also been shown that the cag pathogenicity island remains intact in the coccoid form.[134] Some of these antibiotic resistant cells may remain in the host as persister cells. Following eradication, the persister cells can cause a recurrence of the infection.[132][133] Bacteria can detach from the biofilm to relocate and colonize elsewhere in the stomach to form other biofilms.[90]
Diagnosis
[edit]
Colonization with H. pylori is not a disease in itself, but a condition associated with a number of stomach diseases.[32] Testing is recommended in cases of peptic ulcer disease or low-grade gastric MALT lymphoma; after endoscopic resection of early gastric cancer; for first-degree relatives with gastric cancer, and in certain cases of indigestion. Other indications that prompt testing for H. pylori include long term aspirin or other non-steroidal anti-inflammatory use, unexplained iron deficiency anemia, or in cases of immune thrombocytopenic purpura.[135] Several methods of testing exist, both invasive and non-invasive.
Non-invasive tests for H. pylori infection include serological tests for antibodies, stool tests, and urea breath tests. Carbon urea breath tests include the use of carbon-13, or a radioactive carbon-14 producing a labelled carbon dioxide that can be detected in the breath.[136] Carbon urea breath tests have a high sensitivity and specificity for the diagnosis of H. pylori.[136]
Proton-pump inhibitors and antibiotics should be discontinued for at least 30 days prior to testing for H. pylori infection or eradication, as both agents inhibit H. pylori growth and may lead to false negative results.[135] Testing to confirm eradication is recommended 30 days or more after completion of treatment for H. pylori infection. H. pylori breath testing or stool antigen testing are both reasonable tests to confirm eradication.[135] H. pylori serologic testing, including IgG antibodies, are not recommended as a test of eradication as they may remain elevated for years after successful treatment of infection.[135]
An endoscopic biopsy is an invasive means to test for H. pylori infection. Low-level infections can be missed by biopsy, so multiple samples are recommended. The most accurate method for detecting H. pylori infection is with a histological examination from two sites after endoscopic biopsy, combined with either a rapid urease test or microbial culture.[137] Generally, repeating endoscopy is not recommended to confirm H. pylori eradication, unless there are specific indications to repeat the procedure.[135]
Transmission
[edit]Helicobacter pylori is contagious, and is transmitted through direct contact either with saliva (oral-oral) or feces (fecal–oral route), but mainly through the oral–oral route.[8] Consistent with these transmission routes, the bacteria have been isolated from feces, saliva, and dental plaque.[138] H. pylori may also be transmitted by consuming contaminated food or water.[139] Transmission occurs mainly within families in developed nations, but also from the broader community in developing countries.[140]
Prevention
[edit]To prevent the development of H. pylori-related diseases when infection is suspected, antibiotic-based therapy regimens are recommended to eradicate the bacteria.[46] When successful the disease progression is halted. First line therapy is recommended if low-grade gastric MALT lymphoma is diagnosed, regardless of evidence of H. pylori. However, if a severe condition of atrophic gastritis with gastric lesions is reached antibiotic-based treatment regimens are not advised since such lesions are often not reversible and will progress to gastric cancer.[46] If the cancer is managed to be treated it is advised that an eradication program be followed to prevent a recurrence of infection, or reduce a recurrence of the cancer, known as metachronous.[46][141][142]
Due to H. pylori's role as a major cause of certain diseases (particularly cancers) and its consistently increasing resistance to antibiotic therapy, there is an obvious need for alternative treatments.[143] A vaccine targeted towards the development of gastric cancer, including MALT lymphoma, would also prevent the development of gastric ulcers.[5] A vaccine that would be prophylactic for use in children, and one that would be therapeutic later are the main goals. Challenges to this are the extreme genomic diversity shown by H. pylori and complex host-immune responses.[143][144]
Previous studies in the Netherlands and in the US have shown that such a prophylactic vaccine programme would be ultimately cost-effective.[145][146] However, as of late 2019 there have been no advanced vaccine candidates and only one vaccine in a Phase I clinical trial. Furthermore, development of a vaccine against H. pylori has not been a priority of major pharmaceutical companies.[147] A key target for potential therapy is the proton-gated urea channel, since the secretion of urease enables the survival of the bacterium.[148]
Treatment
[edit]The 2022 Maastricht Consensus Report recognised H. pylori gastritis as Helicobacter pylori induced gastritis, and has been included in ICD11.[44][45][46] Initially the infection tends to be superficial, localised to the upper mucosal layers of the stomach.[149] The intensity of chronic inflammation is related to the cytotoxicity of the H. pylori strain. A greater cytotoxicity will result in the change from a non-atrophic gastritis to an atrophic gastritis, with the loss of mucous glands. This condition is a prequel to the development of peptic ulcers and gastric adenocarcinoma.[149]
Eradication of H. pylori is recommended to treat the infection, including when advanced to peptic ulcer disease. The recommendations for first-line treatment is a quadruple therapy consisting of a proton-pump inhibitor, amoxicillin, clarithromycin, and metronidazole. Prior to treatment, testing is recommended to identify any pre-existing antibiotic resistances. A high rate of resistance to metronidazole has been observed. In areas of known clarithromycin resistance, the first-line therapy is changed to a bismuth based regimen including tetracycline and metronidazole for 14 days. If one of these courses of treatment fails, it is suggested to use the alternative.[44]
Treatment failure may typically be attributed to antibiotic resistance, or inadequate acid suppression from proton-pump inhibitors.[150] Following clinical trials, the use of the potassium-competitive acid blocker vonoprazan, which has a greater acid suppressive action, was approved for use in the US in 2022.[151][150] Its recommended use is in combination with amoxicillin, with or without clarithromycin. It has been shown to have a faster action and can be used with or without food.[150] Successful eradication regimens have revolutionised the treatment of peptic ulcers.[152][153] Eradication of H. pylori is also associated with a subsequent decreased risk of duodenal or gastric ulcer recurrence.[135]
Plant extracts and probiotic foods are being increasingly used as add-ons to usual treatments. Probiotic yogurts containing lactic acid bacteria Bifidobacteria and Lactobacillus exert a suppressive effect on H. pylori infection, and their use has been shown to improve the rates of eradication.[14] Some commensal intestinal bacteria as part of the gut microbiota produce butyrate that acts as a prebiotic and enhances the mucosal immune barrier. Their use as probiotics may help balance the gut dysbiosis that accompanies antibiotic use.[154] Some probiotic strains have been shown to have bactericidal and bacteriostatic activity against H. pylori, and also help to balance the gut dysbiosis.[155][134] Antibiotics have a negative impact on gastrointestinal microbiota and cause nausea, diarrhea, and sickness for which probiotics can alleviate.[134]
Antibiotic resistance
[edit]Increasing antibiotic resistance is the main cause of initial treatment failure. Factors linked to resistance include mutations, efflux pumps, and the formation of biofilms.[156][157] One of the main antibiotics used in eradication therapies is clarithromycin, but clarithromycin-resistant strains have become well-established and the use of alternative antibiotics needs to be considered. Fortunately, non-invasive stool tests for clarithromycin have become available that allow selection of patients that are likely to respond to the therapy.[158] Multidrug resistance has also increased.[157] Additional rounds of antibiotics or other therapies may be used.[159][160][161] Next generation sequencing is looked to for identifying initial specific antibiotic resistances that will help in targeting more effective treatment.[162]
In 2018, the WHO listed H. pylori as a high priority pathogen for the research and discovery of new drugs and treatments.[163] The increasing antibiotic resistance encountered has spurred interest in developing alternative therapies using a number of plant compounds.[164][165] Plant compounds have fewer side effects than synthetic drugs. Most plant extracts contain a complex mix of components that may not act on their own as antimicrobials but can work together with antibiotics to enhance treatment and work towards overcoming resistance.[164] Plant compounds have a different mechanism of action that has proved useful in fighting antimicrobial resistance. For example, various compounds can act by inhibiting enzymes such as urease, and weakening adhesions to the mucous membrane.[166] Sulfur-containing compounds from plants with high concentrations of polysulfides, coumarins, and terpenes have all been shown to be effective against H. pylori.[164]
H. pylori is found in saliva and dental plaque. Its transmission is known to include oral-oral, suggesting that the dental plaque biofilm may act as a reservoir for the bacteria. Periodontal therapy or scaling and root planing has therefore been suggested as an additional treatment to enhance eradication rates, but more research is needed.[139][167]
Cancers
[edit]Stomach cancer
[edit]Helicobacter pylori is a risk factor for gastric adenocarcinomas.[168] Treatment is highly aggressive, with even localized disease being treated sequentially with chemotherapy and radiotherapy before surgical resection.[169] Since this cancer, once developed, is independent of H. pylori infection, eradication regimens are not used.[170]
Gastric MALT lymphoma and DLBCL
[edit]MALT lymphomas are malignancies of mucosa-associated lymphoid tissue. Early gastric MALTomas due to H. pylori may be successfully treated (70–95% of cases) with one or more eradication programs.[14] Some 50–80% of patients who experience eradication of the pathogen develop a remission and long-term clinical control of their lymphoma within 3–28 months. Radiation therapy to the stomach and surrounding (i.e. peri-gastric) lymph nodes has also been used to successfully treat these localized cases. Patients with non-localized (i.e. systemic Ann Arbor stage III and IV) disease who are free of symptoms have been treated with watchful waiting or, if symptomatic, with the immunotherapy drug rituximab (given for 4 weeks) combined with the chemotherapy drug chlorambucil for 6–12 months; 58% of these patients attain a 58% progression-free survival rate at 5 years. Frail stage III/IV patients have been successfully treated with rituximab or the chemotherapy drug cyclophosphamide alone.[171] Antibiotic-proton pump inhibitor eradication therapy and localized radiation therapy have been used successfully to treat H. pylori-positive MALT lymphomas of the rectum; however radiation therapy has given slightly better results and therefore been suggested to be the disease's preferred treatment.[172] However, the generally recognized treatment of choice for patients with systemic involvement uses various chemotherapy drugs often combined with rituximab.
A MALT lymphoma may rarely transform into a more aggressive diffuse large B-cell lymphoma (DLBCL).[173] Where this is associated with H. pylori infection, the DLBCL is less aggressive and more amenable to treatment.[174][175][176] When limited to the stomach, they have sometimes been successfully treated with H. pylori eradication programs.[54][175][177][176] If unresponsive or showing a deterioration, a more conventional chemotherapy (CHOP), immunotherapy, or local radiotherapy can be considered, and any of these or a combination have successfully treated these more advanced types.[175][176]
Prognosis
[edit]Helicobacter pylori colonizes the stomach for decades in most people, and induces chronic gastritis, a long-lasting inflammation of the stomach. In most cases symptoms are never experienced but about 10–20% of those infected will ultimately develop gastric and duodenal ulcers, and have a possible 1–2% lifetime risk of gastric cancer.[65]
H. pylori thrives in a high salt diet, which is seen as an environmental risk factor for its association with gastric cancer. A diet high in salt enhances colonization, increases inflammation, increases the expression of H. pylori virulence factors, and intensifies chronic gastritis.[178][179] Paradoxically, extracts of kimchi, a salted probiotic food, has been found to have a preventive effect on H. pylori–associated gastric carcinogenesis.[180]
In the absence of treatment, H. pylori infection usually persists for life.[181] Infection may disappear in the elderly as the stomach's mucosa becomes increasingly atrophic and inhospitable to colonization. Some studies in young children up to two years of age have shown that infection can be transient in this age group.[182][183]
It is possible for H. pylori to re-establish in a person after eradication. This recurrence can be caused by the original strain (recrudescence), or be caused by a different strain (reinfection). A 2017 meta-analysis showed that the global per-person annual rates of recurrence, reinfection, and recrudescence is 4.3%, 3.1%, and 2.2% respectively. It is unclear what the main risk factors are.[184]
Mounting evidence suggests H. pylori has an important role in protection from some diseases.[16] The incidence of acid reflux disease, Barrett's esophagus, and esophageal cancer have been rising dramatically at the same time as H. pylori's presence decreases.[185] In 1996, Martin J. Blaser advanced the hypothesis that H. pylori has a beneficial effect by regulating the acidity of the stomach contents.[51][185] The hypothesis is not universally accepted, as several randomized controlled trials failed to demonstrate worsening of acid reflux disease symptoms following eradication of H. pylori.[186][187] Nevertheless, Blaser has reasserted his view that H. pylori is a member of the normal gastric microbiota.[17] He postulates that the changes in gastric physiology caused by the loss of H. pylori account for the recent increase in incidence of several diseases, including type 2 diabetes, obesity, and asthma.[17][188] His group has recently shown that H. pylori colonization is associated with a lower incidence of childhood asthma.[189]
Epidemiology
[edit]In 2023, it was estimated that about two-thirds of the world's population were infected with H. pylori infection, being more common in developing countries.[20] H. pylori infection is more prevalent in South America, Sub-Saharan Africa, and the Middle East.[153] The global prevalence declined markedly in the decade following 2010, with a particular reduction in Africa.[21]
The age when someone acquires this bacterium seems to influence the pathologic outcome of the infection. People infected at an early age are likely to develop more intense inflammation that may be followed by atrophic gastritis with a higher subsequent risk of gastric ulcer, gastric cancer, or both. Acquisition at an older age brings different gastric changes more likely to lead to duodenal ulcer.[181] Infections are usually acquired in early childhood in all countries.[32] However, the infection rate of children in developing nations is higher than in industrialized nations, probably due to poor sanitary conditions, perhaps combined with lower antibiotics usage for unrelated pathologies. In developed nations, it is currently uncommon to find infected children, but the percentage of infected people increases with age. The higher prevalence among the elderly reflects higher infection rates incurred in childhood.[32] In the United States, prevalence appears higher in African-American and Hispanic populations, most likely due to socioeconomic factors.[190][191] The lower rate of infection in the West is largely attributed to higher hygiene standards and widespread use of antibiotics. Despite high rates of infection in certain areas of the world, the overall frequency of H. pylori infection is declining.[192] However, antibiotic resistance is appearing in H. pylori; many metronidazole- and clarithromycin-resistant strains are found in most parts of the world.[193]
History
[edit]Helicobacter pylori migrated out of Africa along with its human host around 60,000 years ago.[194] Research has shown that genetic diversity in H. pylori, like that of its host, decreases with geographic distance from East Africa. Using the genetic diversity data, researchers have created simulations that indicate the bacteria seem to have spread from East Africa around 58,000 years ago. Their results indicate modern humans were already infected by H. pylori before their migrations out of Africa, and it has remained associated with human hosts since that time.[195]
H. pylori was first discovered in the stomachs of patients with gastritis and ulcers in 1982 by Barry Marshall and Robin Warren of Perth, Western Australia. At the time, the conventional thinking was that no bacterium could live in the acid environment of the human stomach. In recognition of their discovery, Marshall and Warren were awarded the 2005 Nobel Prize in Physiology or Medicine.[196]
Before the research of Marshall and Warren, German scientists found spiral-shaped bacteria in the lining of the human stomach in 1875, but they were unable to culture them, and the results were eventually forgotten.[185] The Italian researcher Giulio Bizzozero described similarly shaped bacteria living in the acidic environment of the stomach of dogs in 1893.[197] Professor Walery Jaworski of the Jagiellonian University in Kraków investigated sediments of gastric washings obtained by lavage from humans in 1899. Among some rod-like bacteria, he also found bacteria with a characteristic spiral shape, which he called Vibrio rugula. He was the first to suggest a possible role of this organism in the pathogenesis of gastric diseases. His work was included in the Handbook of Gastric Diseases, but it had little impact, as it was published only in Polish.[198] Several small studies conducted in the early 20th century demonstrated the presence of curved rods in the stomachs of many people with peptic ulcers and stomach cancers.[199] Interest in the bacteria waned, however, when an American study published in 1954 failed to observe the bacteria in 1180 stomach biopsies.[200]
Interest in understanding the role of bacteria in stomach diseases was rekindled in the 1970s, with the visualization of bacteria in the stomachs of people with gastric ulcers.[201] The bacteria had also been observed in 1979, by Robin Warren, who researched it further with Barry Marshall from 1981. After unsuccessful attempts at culturing the bacteria from the stomach, they finally succeeded in visualizing colonies in 1982, when they unintentionally left their Petri dishes incubating for five days over the Easter weekend. In their original paper, Warren and Marshall contended that most stomach ulcers and gastritis were caused by bacterial infection and not by stress or spicy food, as had been assumed before.[202]
Some skepticism was expressed initially, but within a few years multiple research groups had verified the association of H. pylori with gastritis and, to a lesser extent, ulcers.[203] To demonstrate H. pylori caused gastritis and was not merely a bystander, Marshall drank a beaker of H. pylori culture. He became ill with nausea and vomiting several days later. An endoscopy 10 days after inoculation revealed signs of gastritis and the presence of H. pylori. These results suggested H. pylori was the causative agent. Marshall and Warren went on to demonstrate antibiotics are effective in the treatment of many cases of gastritis. In 1994, the National Institutes of Health stated most recurrent duodenal and gastric ulcers were caused by H. pylori, and recommended antibiotics be included in the treatment regimen.[204]
The bacterium was initially named Campylobacter pyloridis, then renamed C. pylori in 1987 (pylori being the genitive of pylorus, the circular opening leading from the stomach into the duodenum, from the Ancient Greek word πυλωρός, which means gatekeeper[205]).[206] When 16S ribosomal RNA gene sequencing and other research showed in 1989 that the bacterium did not belong in the genus Campylobacter, it was placed in its own genus, Helicobacter from the Ancient Greek έλιξ (hělix) "spiral" or "coil".[205][207]
In October 1987, a group of experts met in Copenhagen to found the European Helicobacter Study Group (EHSG), an international multidisciplinary research group and the only institution focused on H. pylori.[208] The Group is involved with the Annual International Workshop on Helicobacter and Related Bacteria,[209] (renamed as the European Helicobacter and Microbiota Study Group[210]), the Maastricht Consensus Reports (European Consensus on the management of H. pylori),[211][212][213][214] and other educational and research projects, including two international long-term projects:
- European Registry on H. pylori Management (Hp-EuReg) – a database systematically registering the routine clinical practice of European gastroenterologists.[215]
- Optimal H. pylori management in primary care (OptiCare) – a long-term educational project aiming to disseminate the evidence based recommendations of the Maastricht IV Consensus to primary care physicians in Europe, funded by an educational grant from United European Gastroenterology.[216][217]
Research
[edit]Results from in vitro studies suggest that fatty acids, mainly polyunsaturated fatty acids, have a bactericidal effect against H. pylori, but their in vivo effects have not been proven.[218]
The antibiotic resistance provided by biofilms has generated much research into targeting the mechanisms of quorum sensing used in the formation of biofilms.[88]
A suitable vaccine for H. pylori, either prophylactic or therapeutic, is an ongoing research aim.[8] The Murdoch Children's Research Institute is working at developing a vaccine that instead of specifically targeting the bacteria, aims to inhibit the inflammation caused that leads to the associated diseases.[147]
Gastric organoids can be used as models for the study of H. pylori pathogenesis.[95]
See also
[edit]References
[edit]- ^ a b c Martínez LE, O'Brien VP, Leverich CK, Knoblaugh SE, Salama NR (July 2019). "Nonhelical Helicobacter pylori Mutants Show Altered Gland Colonization and Elicit Less Gastric Pathology than Helical Bacteria during Chronic Infection". Infect Immun. 87 (7). doi:10.1128/IAI.00904-18. PMC 6589060. PMID 31061142.
- ^ a b Salama NR (April 2020). "Cell morphology as a virulence determinant: lessons from Helicobacter pylori". Curr Opin Microbiol. 54: 11–17. doi:10.1016/j.mib.2019.12.002. PMC 7247928. PMID 32014717.
- ^ a b Rust M, Schweinitzer T, Josenhans C (2008). "Helicobacter Flagella, Motility and Chemotaxis". In Yamaoka, Y. (ed.). Helicobacter pylori: Molecular Genetics and Cellular Biology. Caister Academic Press. ISBN 978-1-904455-31-8. Archived from the original on 18 August 2016. Retrieved 1 April 2008.
- ^ Warren JR, Marshall B (June 1983). "Unidentified curved bacilli on gastric epithelium in active chronic gastritis". Lancet. 1 (8336): 1273–5. doi:10.1016/S0140-6736(83)92719-8. PMID 6134060. S2CID 1641856.
- ^ a b FitzGerald R, Smith SM (2021). "An Overview of Helicobacter pylori Infection". Helicobacter Pylori. Methods Mol Biol. Vol. 2283. pp. 1–14. doi:10.1007/978-1-0716-1302-3_1. ISBN 978-1-0716-1301-6. PMID 33765303. S2CID 232365068.
- ^ Watts G (October 2005). "Nobel prize is awarded to doctors who discovered H pylori". BMJ. 331 (7520): 795. doi:10.1136/bmj.331.7520.795. PMC 1246068. PMID 16210262.
- ^ a b "Helicobacter pylori (H. pylori) and Cancer - NCI". www.cancer.gov. 25 September 2013. Archived from the original on 19 October 2023. Retrieved 18 October 2023.
- ^ a b c d e de Brito BB, da Silva FA, Soares AS, Pereira VA, Santos ML, Sampaio MM, et al. (October 2019). "Pathogenesis and clinical management of Helicobacter pylori gastric infection". World J Gastroenterol. 25 (37): 5578–5589. doi:10.3748/wjg.v25.i37.5578. PMC 6785516. PMID 31602159.
- ^ Chen CC, Liou JM, Lee YC, Hong TC, El-Omar EM, Wu MS (2021). "The interplay between Helicobacter pylori and gastrointestinal microbiota". Gut Microbes. 13 (1): 1–22. doi:10.1080/19490976.2021.1909459. PMC 8096336. PMID 33938378.
- ^ a b c d Matsuo Y, Kido Y, Yamaoka Y (March 2017). "Helicobacter pylori Outer Membrane Protein-Related Pathogenesis". Toxins. 9 (3): 101. doi:10.3390/toxins9030101. PMC 5371856. PMID 28287480.
- ^ a b c Marghalani AM, Bin Salman TO, Faqeeh FJ, Asiri MK, Kabel AM (June 2020). "Gastric carcinoma: Insights into risk factors, methods of diagnosis, possible lines of management, and the role of primary care". J Family Med Prim Care. 9 (6): 2659–2663. doi:10.4103/jfmpc.jfmpc_527_20. PMC 7491774. PMID 32984103.
- ^ Koga Y (December 2022). "Microbiota in the stomach and application of probiotics to gastroduodenal diseases". World J Gastroenterol. 28 (47): 6702–6715. doi:10.3748/wjg.v28.i47.6702. PMC 9813937. PMID 36620346.
- ^ a b Shin WS, Xie F, Chen B, Yu J, Lo KW, Tse GM, et al. (October 2023). "Exploring the Microbiome in Gastric Cancer: Assessing Potential Implications and Contextualizing Microorganisms beyond H. pylori and Epstein-Barr Virus". Cancers. 15 (20): 4993. doi:10.3390/cancers15204993. PMC 10605912. PMID 37894360.
- ^ a b c d Violeta Filip P, Cuciureanu D, Sorina Diaconu L, Maria Vladareanu A, Silvia Pop C (2018). "MALT lymphoma: epidemiology, clinical diagnosis and treatment". Journal of Medicine and Life. 11 (3): 187–193. doi:10.25122/jml-2018-0035. PMC 6197515. PMID 30364585.
- ^ Ruggiero P (November 2014). "Use of probiotics in the fight against Helicobacter pylori". World J Gastrointest Pathophysiol. 5 (4): 384–91. doi:10.4291/wjgp.v5.i4.384. PMC 4231502. PMID 25400981.
- ^ a b c d Santos ML, de Brito BB, da Silva FA, Sampaio MM, Marques HS, Oliveira E, et al. (July 2020). "Helicobacter pylori infection: Beyond gastric manifestations". World J Gastroenterol. 26 (28): 4076–4093. doi:10.3748/wjg.v26.i28.4076. PMC 7403793. PMID 32821071.
- ^ a b c Blaser MJ (October 2006). "Who are we? Indigenous microbes and the ecology of human diseases". EMBO Reports. 7 (10): 956–60. doi:10.1038/sj.embor.7400812. PMC 1618379. PMID 17016449.
- ^ Gravina AG, Zagari RM, De Musis C, Romano L, Loguercio C, Romano M (August 2018). "Helicobacter pylori and extragastric diseases: A review". World Journal of Gastroenterology (Review). 24 (29): 3204–3221. doi:10.3748/wjg.v24.i29.3204. PMC 6079286. PMID 30090002.
- ^ Ackerman J (June 2012). "The ultimate social network". Scientific American. Vol. 306, no. 6. pp. 36–43. doi:10.1038/scientificamerican0612-36. PMID 22649992.
- ^ a b "Helicobacter pylori | CDC Yellow Book 2024". wwwnc.cdc.gov. Archived from the original on 22 October 2023. Retrieved 20 October 2023.
- ^ a b Li Y, Choi H, Leung K, Jiang F, Graham DY, Leung WK (19 April 2023). "Global prevalence of Helicobacter pylori infection between 1980 and 2022: a systematic review and meta-analysis". The Lancet Gastroenterology & Hepatology. 8 (6): 553–564. doi:10.1016/S2468-1253(23)00070-5. PMID 37086739. S2CID 258272798.
- ^ Hooi JK, Lai WY, Ng WK, Suen MM, Underwood FE, Tanyingoh D, et al. (August 2017). "Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis". Gastroenterology. 153 (2): 420–429. doi:10.1053/j.gastro.2017.04.022. PMID 28456631.
- ^ Goodwin CS, Armstrong JA, Chilvers T, et al. (1989). "Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov., respectively". Int. J. Syst. Bacteriol. 39 (4): 397–405. doi:10.1099/00207713-39-4-397.
- ^ a b Martínez LE, Hardcastle JM, Wang J, Pincus Z, Tsang J, Hoover TR, et al. (January 2016). "Helicobacter pylori strains vary cell shape and flagellum number to maintain robust motility in viscous environments". Mol Microbiol. 99 (1): 88–110. doi:10.1111/mmi.13218. PMC 4857613. PMID 26365708.
- ^ O'Rourke J, Bode G (2001). Morphology and Ultrastructure. ASM Press. ISBN 978-1-55581-213-3. PMID 21290748.
- ^ a b Kao CY, Sheu BS, Wu JJ (February 2016). "Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis". Biomedical Journal. 39 (1): 14–23. doi:10.1016/j.bj.2015.06.002. PMC 6138426. PMID 27105595.
- ^ Ierardi E, Losurdo G, Mileti A, Paolillo R, Giorgio F, Principi M, et al. (May 2020). "The Puzzle of Coccoid Forms of Helicobacter pylori: Beyond Basic Science". Antibiotics. 9 (6): 293. doi:10.3390/antibiotics9060293. PMC 7345126. PMID 32486473.
- ^ Luo Q, Liu N, Pu S, Zhuang Z, Gong H, Zhang D (2023). "A review on the research progress on non-pharmacological therapy of Helicobacter pylori". Front Microbiol. 14: 1134254. doi:10.3389/fmicb.2023.1134254. PMC 10063898. PMID 37007498.
- ^ Olson JW, Maier RJ (November 2002). "Molecular hydrogen as an energy source for Helicobacter pylori". Science. 298 (5599): 1788–90. Bibcode:2002Sci...298.1788O. doi:10.1126/science.1077123. PMID 12459589. S2CID 27205768.
- ^ a b c Baj J, Forma A, Sitarz M, Portincasa P, Garruti G, Krasowska D, et al. (December 2020). "Helicobacter pylori Virulence Factors-Mechanisms of Bacterial Pathogenicity in the Gastric Microenvironment". Cells. 10 (1): 27. doi:10.3390/cells10010027. PMC 7824444. PMID 33375694.
- ^ Elshenawi Y, Hu S, Hathroubi S (July 2023). "Biofilm of Helicobacter pylori: Life Cycle, Features, and Treatment Options". Antibiotics. 12 (8): 1260. doi:10.3390/antibiotics12081260. PMC 10451559. PMID 37627679.
- ^ a b c d e f g h i j Kusters JG, van Vliet AH, Kuipers EJ (July 2006). "Pathogenesis of Helicobacter pylori infection". Clinical Microbiology Reviews. 19 (3): 449–90. doi:10.1128/CMR.00054-05. PMC 1539101. PMID 16847081.
- ^ a b Alzahrani S, Lina TT, Gonzalez J, Pinchuk IV, Beswick EJ, Reyes VE (September 2014). "Effect of Helicobacter pylori on gastric epithelial cells". World J Gastroenterol. 20 (36): 12767–80. doi:10.3748/wjg.v20.i36.12767. PMC 4177462. PMID 25278677.
- ^ "Genome information for the H. pylori 26695 and J99 strains". Institut Pasteur. 2002. Archived from the original on 26 November 2017. Retrieved 1 September 2008.
- ^ "Helicobacter pylori J99, complete genome". National Center for Biotechnology Information. Archived from the original on 6 April 2011. Retrieved 1 September 2008.
- ^ Oh JD, Kling-Bäckhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, et al. (June 2006). "The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression". Proceedings of the National Academy of Sciences of the United States of America. 103 (26): 9999–10004. Bibcode:2006PNAS..103.9999O. doi:10.1073/pnas.0603784103. PMC 1480403. PMID 16788065.
- ^ "Helicobacter pylori 26695 genome assembly ASM30779v1". NCBI. Retrieved 4 June 2024.
- ^ a b c d Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. (August 1997). "The complete genome sequence of the gastric pathogen Helicobacter pylori". Nature. 388 (6642): 539–47. Bibcode:1997Natur.388..539T. doi:10.1038/41483. PMID 9252185. S2CID 4411220.
- ^ van Vliet AH (January 2017). "Use of pan-genome analysis for the identification of lineage-specific genes of Helicobacter pylori". FEMS Microbiology Letters. 364 (2): fnw296. doi:10.1093/femsle/fnw296. PMID 28011701.
- ^ Uchiyama I, Albritton J, Fukuyo M, Kojima KK, Yahara K, Kobayashi I (9 August 2016). "A Novel Approach to Helicobacter pylori Pan-Genome Analysis for Identification of Genomic Islands". PLOS ONE. 11 (8): e0159419. Bibcode:2016PLoSO..1159419U. doi:10.1371/journal.pone.0159419. PMC 4978471. PMID 27504980.
- ^ a b c Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. (March 2010). "The primary transcriptome of the major human pathogen Helicobacter pylori". Nature. 464 (7286): 250–5. Bibcode:2010Natur.464..250S. doi:10.1038/nature08756. PMID 20164839. S2CID 205219639.
- ^ Müller SA, Pernitzsch SR, Haange SB, Uetz P, von Bergen M, Sharma CM, et al. (3 August 2015). "Stable isotope labeling by amino acids in cell culture based proteomics reveals differences in protein abundances between spiral and coccoid forms of the gastric pathogen Helicobacter pylori". Journal of Proteomics. 126: 34–45. doi:10.1016/j.jprot.2015.05.011. ISSN 1874-3919. PMID 25979772. S2CID 415255. Archived from the original on 27 July 2021. Retrieved 26 July 2021.
- ^ Wuchty S, Müller SA, Caufield JH, Häuser R, Aloy P, Kalkhof S, et al. (May 2018). "Proteome Data Improves Protein Function Prediction in the Interactome of Helicobacter pylori". Mol Cell Proteomics. 17 (5): 961–973. doi:10.1074/mcp.RA117.000474. PMC 5930399. PMID 29414760.
- ^ a b c Malfertheiner P, Megraud F, Rokkas T, Gisbert JP, Liou JM, Schulz C, et al. (August 2022). "Management of Helicobacter pylori infection: the Maastricht VI/Florence consensus report". Gut. 71 (9): 1724–1762. doi:10.1136/gutjnl-2022-327745. hdl:10486/714546. PMID 35944925.
- ^ a b "ICD-11 for Mortality and Morbidity Statistics". icd.who.int. Archived from the original on 15 October 2023. Retrieved 9 January 2024.
- ^ a b c d e "The Changes Made in the New Expert Consensus on H pylori". Medscape. Archived from the original on 9 January 2024. Retrieved 9 January 2024.
- ^ Repetto O, Vettori R, Steffan A, Cannizzaro R, De Re V (November 2023). "Circulating Proteins as Diagnostic Markers in Gastric Cancer". Int J Mol Sci. 24 (23): 16931. doi:10.3390/ijms242316931. PMC 10706891. PMID 38069253.
- ^ Livzan MA, Mozgovoi SI, Gaus OV, Shimanskaya AG, Kononov AV (July 2023). "Histopathological Evaluation of Gastric Mucosal Atrophy for Predicting Gastric Cancer Risk: Problems and Solutions". Diagnostics. 13 (15): 2478. doi:10.3390/diagnostics13152478. PMC 10417051. PMID 37568841.
- ^ Dixon MF (February 2000). "Patterns of inflammation linked to ulcer disease". Baillière's Best Practice & Research. Clinical Gastroenterology. 14 (1): 27–40. doi:10.1053/bega.1999.0057. PMID 10749087.
- ^ Mommersteeg MC, Yu BT, van den Bosch TP, von der Thüsen JH, Kuipers EJ, Doukas M, et al. (October 2022). "Constitutive programmed death ligand 1 expression protects gastric G-cells from Helicobacter pylori-induced inflammation". Helicobacter. 27 (5): e12917. doi:10.1111/hel.12917. PMC 9542424. PMID 35899973. S2CID 251132578.
- ^ a b Blaser MJ, Atherton JC (February 2004). "Helicobacter pylori persistence: biology and disease". The Journal of Clinical Investigation. 113 (3): 321–33. doi:10.1172/JCI20925. PMC 324548. PMID 14755326.
- ^ Schubert ML, Peura DA (June 2008). "Control of gastric acid secretion in health and disease". Gastroenterology. 134 (7): 1842–60. doi:10.1053/j.gastro.2008.05.021. PMID 18474247. S2CID 206210451.
- ^ Abbas H, Niazi M, Makker J (May 2017). "Mucosa-Associated Lymphoid Tissue (MALT) Lymphoma of the Colon: A Case Report and a Literature Review". The American Journal of Case Reports. 18: 491–497. doi:10.12659/AJCR.902843. PMC 5424574. PMID 28469125.
- ^ a b Paydas S (April 2015). "Helicobacter pylori eradication in gastric diffuse large B cell lymphoma". World Journal of Gastroenterology. 21 (13): 3773–6. doi:10.3748/wjg.v21.i13.3773. PMC 4385524. PMID 25852262.
- ^ Kuipers EJ (March 1999). "Review article: exploring the link between Helicobacter pylori and gastric cancer". Alimentary Pharmacology & Therapeutics. 13 (Suppl 1): 3–11. doi:10.1046/j.1365-2036.1999.00002.x. PMID 10209681. S2CID 19231673.
- ^ Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. (April 2019). "Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods". International Journal of Cancer. 144 (8): 1941–1953. doi:10.1002/ijc.31937. PMID 30350310.
- ^ Deng JY, Liang H (April 2014). "Clinical significance of lymph node metastasis in gastric cancer". World Journal of Gastroenterology. 20 (14): 3967–75. doi:10.3748/wjg.v20.i14.3967. PMC 3983452. PMID 24744586.
- ^ a b Valenzuela MA, Canales J, Corvalán AH, Quest AF (December 2015). "Helicobacter pylori-induced inflammation and epigenetic changes during gastric carcinogenesis". World Journal of Gastroenterology. 21 (45): 12742–56. doi:10.3748/wjg.v21.i45.12742. PMC 4671030. PMID 26668499.
- ^ a b Raza Y, Khan A, Farooqui A, Mubarak M, Facista A, Akhtar SS, et al. (October 2014). "Oxidative DNA damage as a potential early biomarker of Helicobacter pylori associated carcinogenesis". Pathology & Oncology Research. 20 (4): 839–46. doi:10.1007/s12253-014-9762-1. PMID 24664859. S2CID 18727504.
- ^ Koeppel M, Garcia-Alcalde F, Glowinski F, Schlaermann P, Meyer TF (June 2015). "Helicobacter pylori Infection Causes Characteristic DNA Damage Patterns in Human Cells". Cell Reports. 11 (11): 1703–13. doi:10.1016/j.celrep.2015.05.030. PMID 26074077.
- ^ a b c d Markowski AR, Markowska A, Guzinska-Ustymowicz K (October 2016). "Pathophysiological and clinical aspects of gastric hyperplastic polyps". World Journal of Gastroenterology. 22 (40): 8883–8891. doi:10.3748/wjg.v22.i40.8883. PMC 5083793. PMID 27833379.
- ^ Dong YF, Guo T, Yang H, Qian JM, Li JN (February 2019). "[Correlations between gastric Helicobacter pylori infection and colorectal polyps or cancer]". Zhonghua Nei Ke Za Zhi (in Chinese). 58 (2): 139–142. doi:10.3760/cma.j.issn.0578-1426.2019.02.011. PMID 30704201.
- ^ a b Zuo Y, Jing Z, Bie M, Xu C, Hao X, Wang B (September 2020). "Association between Helicobacter pylori infection and the risk of colorectal cancer: A systematic review and meta-analysis". Medicine (Baltimore). 99 (37): e21832. doi:10.1097/MD.0000000000021832. PMC 7489651. PMID 32925719.
- ^ Papastergiou V, Karatapanis S, Georgopoulos SD (January 2016). "Helicobacter pylori and colorectal neoplasia: Is there a causal link?". World J Gastroenterol. 22 (2): 649–58. doi:10.3748/wjg.v22.i2.649. PMC 4716066. PMID 26811614.
- ^ a b c Debowski AW, Walton SM, Chua EG, Tay AC, Liao T, Lamichhane B, et al. (June 2017). "Helicobacter pylori gene silencing in vivo demonstrates urease is essential for chronic infection". PLOS Pathogens. 13 (6): e1006464. doi:10.1371/journal.ppat.1006464. PMC 5500380. PMID 28644872.
- ^ Al Quraan AM, Beriwal N, Sangay P, Namgyal T (October 2019). "The Psychotic Impact of Helicobacter pylori Gastritis and Functional Dyspepsia on Depression: A Systematic Review". Cureus. 11 (10): e5956. doi:10.7759/cureus.5956. PMC 6863582. PMID 31799095.
- ^ a b "Helicobacter Pylori (H. Pylori) Tests: MedlinePlus Medical Test". medlineplus.gov. Archived from the original on 16 February 2024. Retrieved 16 February 2024.
- ^ a b "Symptoms & Causes of Peptic Ulcers (Stomach or Duodenal Ulcers) - NIDDK". National Institute of Diabetes and Digestive and Kidney Diseases. Archived from the original on 17 February 2024. Retrieved 17 February 2024.
- ^ Popa DG, Obleagă CV, Socea B, Serban D, Ciurea ME, Diaconescu M, et al. (October 2021). "Role of Helicobacter pylori in the triggering and evolution of hemorrhagic gastro-duodenal lesions". Exp Ther Med. 22 (4): 1147. doi:10.3892/etm.2021.10582. PMC 8392874. PMID 34504592.
- ^ Al-Azri M, Al-Kindi J, Al-Harthi T, Al-Dahri M, Panchatcharam SM, Al-Maniri A (June 2019). "Awareness of Stomach and Colorectal Cancer Risk Factors, Symptoms and Time Taken to Seek Medical Help Among Public Attending Primary Care Setting in Muscat Governorate, Oman". Journal of Cancer Education. 34 (3): 423–434. doi:10.1007/s13187-017-1266-8. ISSN 0885-8195. PMID 28782080. S2CID 4017466. Archived from the original on 24 February 2024. Retrieved 20 January 2024.
- ^ Wu Q, Yang ZP, Xu P, Gao LC, Fan DM (July 2013). "Association between Helicobacter pylori infection and the risk of colorectal neoplasia: a systematic review and meta-analysis". Colorectal Disease. 15 (7): e352-64. doi:10.1111/codi.12284. PMID 23672575. S2CID 5444584.
- ^ Soetikno RM, Kaltenbach T, Rouse RV, Park W, Maheshwari A, Sato T, et al. (March 2008). "Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults". JAMA. 299 (9): 1027–35. doi:10.1001/jama.299.9.1027. PMID 18319413.
- ^ a b Yamaoka Y, Saruuljavkhlan B, Alfaray RI, Linz B (2023). "Pathogenomics of Helicobacter pylori". Helicobacter pylori and Gastric Cancer. Current Topics in Microbiology and Immunology. Vol. 444. pp. 117–155. doi:10.1007/978-3-031-47331-9_5. ISBN 978-3-031-47330-2. PMID 38231217.
- ^ Alfarouk KO, Bashir AH, Aljarbou AN, Ramadan AM, Muddathir AK, AlHoufie ST, et al. (22 February 2019). "Helicobacter pylori in Gastric Cancer and Its Management". Frontiers in Oncology. 9: 75. doi:10.3389/fonc.2019.00075. PMC 6395443. PMID 30854333.
- ^ Santos JC, Ribeiro ML (August 2015). "Epigenetic regulation of DNA repair machinery in Helicobacter pylori-induced gastric carcinogenesis". World Journal of Gastroenterology. 21 (30): 9021–37. doi:10.3748/wjg.v21.i30.9021. PMC 4533035. PMID 26290630.
- ^ Raza Y, Ahmed A, Khan A, Chishti AA, Akhter SS, Mubarak M, et al. (May 2020). "Helicobacter pylori severely reduces expression of DNA repair proteins PMS2 and ERCC1 in gastritis and gastric cancer". DNA Repair. 89: 102836. doi:10.1016/j.dnarep.2020.102836. PMID 32143126.
- ^ Dore MP, Pes GM, Bassotti G, Usai-Satta P (2016). "Dyspepsia: When and How to Test for Helicobacter pylori Infection". Gastroenterology Research and Practice. 2016: 8463614. doi:10.1155/2016/8463614. PMC 4864555. PMID 27239194.
- ^ a b Muhammad JS, Eladl MA, Khoder G (February 2019). "Helicobacter pylori-induced DNA Methylation as an Epigenetic Modulator of Gastric Cancer: Recent Outcomes and Future Direction". Pathogens. 8 (1): 23. doi:10.3390/pathogens8010023. PMC 6471032. PMID 30781778.
- ^ a b Noto JM, Peek RM (2011). "The role of microRNAs in Helicobacter pylori pathogenesis and gastric carcinogenesis". Frontiers in Cellular and Infection Microbiology. 1: 21. doi:10.3389/fcimb.2011.00021. PMC 3417373. PMID 22919587.
- ^ Tsuji S, Kawai N, Tsujii M, Kawano S, Hori M (July 2003). "Review article: inflammation-related promotion of gastrointestinal carcinogenesis--a perigenetic pathway". Alimentary Pharmacology & Therapeutics. 18 (Suppl 1): 82–9. doi:10.1046/j.1365-2036.18.s1.22.x. PMID 12925144. S2CID 22646916.
- ^ Yu B, de Vos D, Guo X, Peng S, Xie W, Peppelenbosch MP, et al. (April 2024). "IL-6 facilitates cross-talk between epithelial cells and tumor- associated macrophages in Helicobacter pylori-linked gastric carcinogenesis". Neoplasia. 50: 100981. doi:10.1016/j.neo.2024.100981. PMC 10912637. PMID 38422751.
- ^ a b Suganuma M, Yamaguchi K, Ono Y, Matsumoto H, Hayashi T, Ogawa T, et al. (July 2008). "TNF-alpha-inducing protein, a carcinogenic factor secreted from H. pylori, enters gastric cancer cells". International Journal of Cancer. 123 (1): 117–22. doi:10.1002/ijc.23484. PMID 18412243. S2CID 5532769.
- ^ Duan Q, Zhou M, Zhu L, Zhu G (January 2013). "Flagella and bacterial pathogenicity". J Basic Microbiol. 53 (1): 1–8. doi:10.1002/jobm.201100335. PMID 22359233. S2CID 22002199.
- ^ a b c Nedeljković M, Sastre DE, Sundberg EJ (July 2021). "Bacterial Flagellar Filament: A Supramolecular Multifunctional Nanostructure". Int J Mol Sci. 22 (14): 7521. doi:10.3390/ijms22147521. PMC 8306008. PMID 34299141.
- ^ Elbehiry A, Marzouk E, Aldubaib M, Abalkhail A, Anagreyyah S, Anajirih N, et al. (January 2023). "Helicobacter pylori Infection: Current Status and Future Prospects on Diagnostic, Therapeutic and Control Challenges". Antibiotics. 12 (2): 191. doi:10.3390/antibiotics12020191. PMC 9952126. PMID 36830102.
- ^ Petersen AM, Krogfelt KA (May 2003). "Helicobacter pylori: an invading microorganism? A review". FEMS Immunology and Medical Microbiology (Review). 36 (3): 117–26. doi:10.1016/S0928-8244(03)00020-8. PMID 12738380.
- ^ Ali A, AlHussaini KI (January 2024). "Helicobacter pylori: A Contemporary Perspective on Pathogenesis, Diagnosis and Treatment Strategies". Microorganisms. 12 (1): 222. doi:10.3390/microorganisms12010222. PMC 10818838. PMID 38276207.
- ^ a b Zafer MM, Mohamed GA, Ibrahim SR, Ghosh S, Bornman C, Elfaky MA (February 2024). "Biofilm-mediated infections by multidrug-resistant microbes: a comprehensive exploration and forward perspectives". Arch Microbiol. 206 (3): 101. Bibcode:2024ArMic.206..101Z. doi:10.1007/s00203-023-03826-z. PMC 10867068. PMID 38353831.
- ^ a b Sun Q, Yuan C, Zhou S, Lu J, Zeng M, Cai X, et al. (2023). "Helicobacter pylori infection: a dynamic process from diagnosis to treatment". Front Cell Infect Microbiol. 13: 1257817. doi:10.3389/fcimb.2023.1257817. PMC 10621068. PMID 37928189.
- ^ a b c d e f Lin Q, Lin S, Fan Z, Liu J, Ye D, Guo P (May 2024). "A Review of the Mechanisms of Bacterial Colonization of the Mammal Gut". Microorganisms. 12 (5): 1026. doi:10.3390/microorganisms12051026. PMC 11124445. PMID 38792855.
- ^ a b c Hernández VM, Arteaga A, Dunn MF (November 2021). "Diversity, properties and functions of bacterial arginases". FEMS Microbiol Rev. 45 (6). doi:10.1093/femsre/fuab034. PMID 34160574.
- ^ a b Li S, Zhao W, Xia L, Kong L, Yang L (2023). "How Long Will It Take to Launch an Effective Helicobacter pylori Vaccine for Humans?". Infect Drug Resist. 16: 3787–3805. doi:10.2147/IDR.S412361. PMC 10278649. PMID 37342435.
- ^ George G, Kombrabail M, Raninga N, Sau AK (March 2017). "Arginase of Helicobacter Gastric Pathogens Uses a Unique Set of Non-catalytic Residues for Catalysis". Biophysical Journal. 112 (6): 1120–1134. Bibcode:2017BpJ...112.1120G. doi:10.1016/j.bpj.2017.02.009. PMC 5376119. PMID 28355540.
- ^ Smoot DT (December 1997). "How does Helicobacter pylori cause mucosal damage? Direct mechanisms". Gastroenterology. 113 (6 Suppl): S31-4, discussion S50. doi:10.1016/S0016-5085(97)80008-X. PMID 9394757.
- ^ a b c Doohan D, Rezkitha YA, Waskito LA, Yamaoka Y, Miftahussurur M (July 2021). "Helicobacter pylori BabA-SabA Key Roles in the Adherence Phase: The Synergic Mechanism for Successful Colonization and Disease Development". Toxins. 13 (7): 485. doi:10.3390/toxins13070485. PMC 8310295. PMID 34357957.
- ^ Rad R, Gerhard M, Lang R, Schöniger M, Rösch T, Schepp W, et al. (15 March 2002). "The Helicobacter pylori Blood Group Antigen-Binding Adhesin Facilitates Bacterial Colonization and Augments a Nonspecific Immune Response". The Journal of Immunology. 168 (6): 3033–3041. doi:10.4049/jimmunol.168.6.3033. PMID 11884476.
- ^ Bugaytsova JA, Björnham O, Chernov YA, Gideonsson P, Henriksson S, Mendez M, et al. (March 2017). "Helicobacter pylori Adapts to Chronic Infection and Gastric Disease via pH-Responsive BabA-Mediated Adherence". Cell Host & Microbe. 21 (3): 376–389. doi:10.1016/j.chom.2017.02.013. PMC 5392239. PMID 28279347.
- ^ Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, Roche N, et al. (July 2002). "Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation". Science. 297 (5581): 573–8. Bibcode:2002Sci...297..573M. doi:10.1126/science.1069076. PMC 2570540. PMID 12142529.
- ^ a b c Testerman TL, Morris J (September 2014). "Beyond the stomach: an updated view of Helicobacter pylori pathogenesis, diagnosis, and treatment". World Journal of Gastroenterology (Review). 20 (36): 12781–808. doi:10.3748/wjg.v20.i36.12781. PMC 4177463. PMID 25278678.
- ^ Zhang L, Xie J (September 2023). "Biosynthesis, structure and biological function of cholesterol glucoside in Helicobacter pylori: A review". Medicine (Baltimore). 102 (36): e34911. doi:10.1097/MD.0000000000034911. PMC 10489377. PMID 37682174.
- ^ Ridyard KE, Overhage J (May 2021). "The Potential of Human Peptide LL-37 as an Antimicrobial and Anti-Biofilm Agent". Antibiotics. 10 (6): 650. doi:10.3390/antibiotics10060650. PMC 8227053. PMID 34072318.
- ^ a b Hsu CY, Yeh JY, Chen CY, Wu HY, Chiang MH, Wu CL, et al. (December 2021). "Helicobacter pylori cholesterol-α-glucosyltransferase manipulates cholesterol for bacterial adherence to gastric epithelial cells". Virulence. 12 (1): 2341–2351. doi:10.1080/21505594.2021.1969171. PMC 8437457. PMID 34506250.
- ^ Morey P, Pfannkuch L, Pang E, Boccellato F, Sigal M, Imai-Matsushima A, et al. (April 2018). "Helicobacter pylori Depletes Cholesterol in Gastric Glands to Prevent Interferon Gamma Signaling and Escape the Inflammatory Response". Gastroenterology. 154 (5): 1391–1404.e9. doi:10.1053/j.gastro.2017.12.008. hdl:21.11116/0000-0001-3B12-9. PMID 29273450.
- ^ a b Ramarao N, Gray-Owen SD, Meyer TF (October 2000). "Helicobacter pylori induces but survives the extracellular release of oxygen radicals from professional phagocytes using its catalase activity". Mol Microbiol. 38 (1): 103–13. doi:10.1046/j.1365-2958.2000.02114.x. hdl:11858/00-001M-0000-000E-C7AD-8. PMID 11029693.
- ^ "UniProt". www.uniprot.org. Retrieved 20 March 2024.
- ^ "TNF-alpha inducing protein". www.uniprot.org. Retrieved 8 April 2024.
- ^ Watanabe T, Takahashi A, Suzuki K, Kurusu-Kanno M, Yamaguchi K, Fujiki H, et al. (15 May 2014). "Epithelial-mesenchymal transition in human gastric cancer cell lines induced by TNF-α-inducing protein of Helicobacter pylori: Cell migration induced by Tipα of H. pylori". International Journal of Cancer. 134 (10): 2373–2382. doi:10.1002/ijc.28582. PMID 24249671.
- ^ Wallden K, Rivera-Calzada A, Waksman G (September 2010). "Type IV secretion systems: versatility and diversity in function". Cell Microbiol. 12 (9): 1203–12. doi:10.1111/j.1462-5822.2010.01499.x. PMC 3070162. PMID 20642798.
- ^ a b Broutet N, Marais A, Lamouliatte H, de Mascarel A, Samoyeau R, Salamon R, et al. (April 2001). "cagA Status and eradication treatment outcome of anti-Helicobacter pylori triple therapies in patients with nonulcer dyspepsia". Journal of Clinical Microbiology. 39 (4): 1319–22. doi:10.1128/JCM.39.4.1319-1322.2001. PMC 87932. PMID 11283049.
- ^ Zawilak-Pawlik A, Zarzecka U, Żyła-Uklejewicz D, Lach J, Strapagiel D, Tegtmeyer N, et al. (August 2019). "Establishment of serine protease htrA mutants in Helicobacter pylori is associated with secA mutations". Scientific Reports. 9 (1): 11794. Bibcode:2019NatSR...911794Z. doi:10.1038/s41598-019-48030-6. PMC 6692382. PMID 31409845.
- ^ Peek RM, Crabtree JE (January 2006). "Helicobacter infection and gastric neoplasia". The Journal of Pathology. 208 (2): 233–48. doi:10.1002/path.1868. PMID 16362989. S2CID 31718278.
- ^ Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, Moran AP, et al. (November 2004). "Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island". Nature Immunology. 5 (11): 1166–74. doi:10.1038/ni1131. PMID 15489856. S2CID 2898805.
- ^ Backert S, Selbach M (August 2008). "Role of type IV secretion in Helicobacter pylori pathogenesis". Cellular Microbiology. 10 (8): 1573–81. doi:10.1111/j.1462-5822.2008.01156.x. PMID 18410539. S2CID 37626.
- ^ Hatakeyama M (September 2004). "Oncogenic mechanisms of the Helicobacter pylori CagA protein". Nature Reviews. Cancer. 4 (9): 688–94. doi:10.1038/nrc1433. PMID 15343275. S2CID 1218835.
- ^ Kim W, Moss SF (2008). "The role of Helicobacter pylori in the pathogenesis of gastric malignancies". Oncology Reviews. 2 (3): 131–140. doi:10.1007/s12156-008-0068-y.
- ^ "UniProt". www.uniprot.org. Retrieved 21 March 2024.
- ^ Miehlke S, Yu J, Schuppler M, Frings C, Kirsch C, Negraszus N, et al. (April 2001). "Helicobacter pylori vacA, iceA, and cagA status and pattern of gastritis in patients with malignant and benign gastroduodenal disease". The American Journal of Gastroenterology. 96 (4): 1008–13. doi:10.1111/j.1572-0241.2001.03685.x. PMID 11316139. S2CID 24024542. Archived from the original on 23 February 2022. Retrieved 24 June 2020.
- ^ a b Hisatsune J, Yamasaki E, Nakayama M, Shirasaka D, Kurazono H, Katagata Y, et al. (September 2007). "Helicobacter pylori VacA enhances prostaglandin E2 production through induction of cyclooxygenase 2 expression via a p38 mitogen-activated protein kinase/activating transcription factor 2 cascade in AZ-521 cells". Infect Immun. 75 (9): 4472–81. doi:10.1128/IAI.00500-07. PMC 1951161. PMID 17591797.
- ^ Capurro MI, Greenfield LK, Prashar A, Xia S, Abdullah M, Wong H, et al. (August 2019). "VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1". Nature Microbiology. 4 (8): 1411–1423. doi:10.1038/s41564-019-0441-6. PMC 6938649. PMID 31110360.
- ^ Sajib S, Zahra FT, Lionakis MS, German NA, Mikelis CM (February 2018). "Mechanisms of angiogenesis in microbe-regulated inflammatory and neoplastic conditions". Angiogenesis. 21 (1): 1–14. doi:10.1007/s10456-017-9583-4. PMID 29110215. S2CID 3346742.
- ^ da Costa DM, Pereira Edos S, Rabenhorst SH (October 2015). "What exists beyond cagA and vacA? Helicobacter pylori genes in gastric diseases". World J Gastroenterol. 21 (37): 10563–72. doi:10.3748/wjg.v21.i37.10563. PMC 4588078. PMID 26457016.
- ^ Dumrese C, Slomianka L, Ziegler U, Choi SS, Kalia A, Fulurija A, et al. (May 2009). "The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype". FEBS Letters. 583 (10): 1637–43. Bibcode:2009FEBSL.583.1637D. doi:10.1016/j.febslet.2009.04.027. PMC 2764743. PMID 19393649.
- ^ Alam J, Sarkar A, Karmakar BC, Ganguly M, Paul S, Mukhopadhyay AK (August 2020). "Novel virulence factor dupA of Helicobacter pylori as an important risk determinant for disease manifestation: An overview". World J Gastroenterol. 26 (32): 4739–4752. doi:10.3748/wjg.v26.i32.4739. PMC 7459207. PMID 32921954.
- ^ a b c d Trastoy R, Manso T, Fernández-García L, Blasco L, Ambroa A, Pérez Del Molino ML, et al. (October 2018). "Mechanisms of Bacterial Tolerance and Persistence in the Gastrointestinal and Respiratory Environments". Clin Microbiol Rev. 31 (4). doi:10.1128/CMR.00023-18. PMC 6148185. PMID 30068737.
- ^ Olczak AA, Olson JW, Maier RJ (June 2002). "Oxidative-stress resistance mutants of Helicobacter pylori". Journal of Bacteriology. 184 (12): 3186–93. doi:10.1128/JB.184.12.3186-3193.2002. PMC 135082. PMID 12029034.
- ^ a b c d Dorer MS, Fero J, Salama NR (July 2010). Blanke SR (ed.). "DNA damage triggers genetic exchange in Helicobacter pylori". PLOS Pathogens. 6 (7): e1001026. doi:10.1371/journal.ppat.1001026. PMC 2912397. PMID 20686662.
- ^ O'Rourke EJ, Chevalier C, Pinto AV, Thiberge JM, Ielpi L, Labigne A, et al. (March 2003). "Pathogen DNA as target for host-generated oxidative stress: role for repair of bacterial DNA damage in Helicobacter pylori colonization". Proceedings of the National Academy of Sciences of the United States of America. 100 (5): 2789–94. Bibcode:2003PNAS..100.2789O. doi:10.1073/pnas.0337641100. PMC 151419. PMID 12601164.
- ^ Michod RE, Bernstein H, Nedelcu AM (May 2008). "Adaptive value of sex in microbial pathogens". Infection, Genetics and Evolution. 8 (3): 267–85. Bibcode:2008InfGE...8..267M. doi:10.1016/j.meegid.2008.01.002. PMID 18295550.
- ^ Ailloud F, Didelot X, Woltemate S, Pfaffinger G, Overmann J, Bader RC, et al. (May 2019). "Within-host evolution of Helicobacter pylori shaped by niche-specific adaptation, intragastric migrations and selective sweeps". Nat Commun. 10 (1): 2273. Bibcode:2019NatCo..10.2273A. doi:10.1038/s41467-019-10050-1. PMC 6531487. PMID 31118420.
- ^ Loughlin MF, Barnard FM, Jenkins D, Sharples GJ, Jenks PJ (April 2003). "Helicobacter pylori mutants defective in RuvC Holliday junction resolvase display reduced macrophage survival and spontaneous clearance from the murine gastric mucosa". Infection and Immunity. 71 (4): 2022–31. doi:10.1128/IAI.71.4.2022-2031.2003. PMC 152077. PMID 12654822.
- ^ a b Wang G, Maier RJ (January 2008). "Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori". Infection and Immunity. 76 (1): 153–60. doi:10.1128/IAI.00791-07. PMC 2223656. PMID 17954726.
- ^ a b Bahmaninejad P, Ghafourian S, Mahmoudi M, Maleki A, Sadeghifard N, Badakhsh B (April 2021). "Persister cells as a possible cause of antibiotic therapy failure in Helicobacter pylori". JGH Open. 5 (4): 493–497. doi:10.1002/jgh3.12527. PMC 8035453. PMID 33860100.
- ^ a b Cammarota G, Sanguinetti M, Gallo A, Posteraro B (August 2012). "Review article: biofilm formation by H elicobacter pylori as a target for eradication of resistant infection". Alimentary Pharmacology & Therapeutics. 36 (3): 222–230. doi:10.1111/j.1365-2036.2012.05165.x. PMID 22650647. S2CID 24026187. Retrieved 3 March 2024.
- ^ a b c d Shadvar N, Akrami S, Mousavi Sagharchi SM, Askandar RH, Merati A, Aghayari M, et al. (7 May 2024). "A review for non-antibiotic treatment of Helicobacter pylori: new insight". Frontiers in Microbiology. 15. doi:10.3389/fmicb.2024.1379209. ISSN 1664-302X. PMC 11106852. PMID 38774508.
- ^ a b c d e f Crowe SE (21 March 2019). "Helicobacter pylori Infection". New England Journal of Medicine. 380 (12): 1158–1165. doi:10.1056/NEJMcp1710945. PMID 30893536. S2CID 84843669.
- ^ a b Jambi LK (7 October 2022). "Systematic Review and Meta-Analysis on the Sensitivity and Specificity of (13)C/(14)C-Urea Breath Tests in the Diagnosis of Helicobacter pylori Infection". Diagnostics. 12 (10): 2428. doi:10.3390/diagnostics12102428. PMC 9600925. PMID 36292117.
- ^ Logan RP, Walker MM (October 2001). "ABC of the upper gastrointestinal tract: Epidemiology and diagnosis of Helicobacter pylori infection". BMJ. 323 (7318): 920–2. doi:10.1136/bmj.323.7318.920. PMC 1121445. PMID 11668141.
- ^ Reshetnyak VI, Burmistrov AI, Maev IV (February 2021). "Helicobacter pylori: Commensal, symbiont or pathogen?". World J Gastroenterol. 27 (7): 545–560. doi:10.3748/wjg.v27.i7.545. PMC 7901052. PMID 33642828.
- ^ a b Rahat M, Saqib M, Ahmed M, Suleman M, Ismail SM, Mumtaz H, et al. (June 2023). "Use of eradication therapy in adjunction to periodontal therapy versus alone for treatment of Helicobacter pylori infections: a mini review". Ann Med Surg (Lond). 85 (6): 2756–2760. doi:10.1097/MS9.0000000000000741. PMC 10289787. PMID 37363585.
- ^ Delport W, van der Merwe SW (2007). "The transmission of Helicobacter pylori: the effects of analysis method and study population on inference". Best Practice & Research. Clinical Gastroenterology. 21 (2): 215–36. doi:10.1016/j.bpg.2006.10.001. hdl:2263/4083. PMID 17382274.
- ^ Tsukamoto T, Nakagawa M, Kiriyama Y, Toyoda T, Cao X (August 2017). "Prevention of Gastric Cancer: Eradication of Helicobacter Pylori and Beyond". International Journal of Molecular Sciences. 18 (8): 1699. doi:10.3390/ijms18081699. PMC 5578089. PMID 28771198.
- ^ Li L, Yu C (2019). "Helicobacter pylori Infection following Endoscopic Resection of Early Gastric Cancer". BioMed Research International. 2019. doi:10.1155/2019/9824964. PMC 6816031. PMID 31737682.
- ^ a b Talebi Bezmin Abadi A (March 2016). "Vaccine against Helicobacter pylori: Inevitable approach". World J Gastroenterol. 22 (11): 3150–7. doi:10.3748/wjg.v22.i11.3150. PMC 4789989. PMID 27003991.
- ^ Blanchard TG, Nedrud JG (2010). "9. Helicobacter pylori Vaccines". In Sutton P, Mitchell H (eds.). Helicobacter pylori in the 21st Century. CABI. pp. 167–189. ISBN 978-1-84593-594-8. Retrieved 7 August 2013.
- ^ de Vries R, Klok RM, Brouwers JR, Postma MJ (February 2009). "Cost-effectiveness of a potential future Helicobacter pylori vaccine in the Netherlands: the impact of varying the discount rate for health". Vaccine. 27 (6): 846–52. doi:10.1016/j.vaccine.2008.11.081. PMID 19084566. Archived from the original on 10 May 2021. Retrieved 7 August 2013.
- ^ Rupnow MF, Chang AH, Shachter RD, Owens DK, Parsonnet J (October 2009). "Cost-effectiveness of a potential prophylactic Helicobacter pylori vaccine in the United States". The Journal of Infectious Diseases. 200 (8): 1311–7. doi:10.1086/605845. PMID 19751153.
- ^ a b Sutton P, Boag JM (November 2019). "Status of vaccine research and development for Helicobacter pylori". Vaccine. 37 (50): 7295–7299. doi:10.1016/j.vaccine.2018.01.001. PMC 6892279. PMID 29627231.
- ^ "PDB101: Molecule of the Month: Proton-Gated Urea Channel". RCSB: PDB-101. Archived from the original on 6 November 2023. Retrieved 6 November 2023.
- ^ a b Azer SA, Akhondi H (2019). "Gastritis". StatPearls. PMID 31334970.
- ^ a b c Shirley M (March 2024). "Vonoprazan: A Review in Helicobacter pylori Infection". Drugs. 84 (3): 319–327. doi:10.1007/s40265-023-01991-5. PMC 11090951. PMID 38388872.
- ^ "Two New Regimens Win FDA Approval for H. Pylori Infection". www.medpagetoday.com. 4 May 2022. Archived from the original on 25 March 2023. Retrieved 25 March 2023.
- ^ Rauws EA, Tytgat GN (May 1990). "Cure of duodenal ulcer associated with eradication of Helicobacter pylori". Lancet. 335 (8700): 1233–5. doi:10.1016/0140-6736(90)91301-P. PMID 1971318. S2CID 41888935.
- ^ a b Burkitt MD, Duckworth CA, Williams JM, Pritchard DM (February 2017). "Helicobacter pylori-induced gastric pathology: insights from in vivo and ex vivo models". Disease Models & Mechanisms. 10 (2): 89–104. doi:10.1242/dmm.027649. PMC 5312008. PMID 28151409.
- ^ Zhu LB, Zhang YC, Huang HH, Lin J (September 2021). "Prospects for clinical applications of butyrate-producing bacteria". World J Clin Pediatr. 10 (5): 84–92. doi:10.5409/wjcp.v10.i5.84. PMC 8465514. PMID 34616650.
- ^ Saracino IM, Pavoni M, Saccomanno L, Fiorini G, Pesci V, Foschi C, et al. (May 2020). "Antimicrobial Efficacy of Five Probiotic Strains Against Helicobacter pylori". Antibiotics. 9 (5): 244. doi:10.3390/antibiotics9050244. PMC 7277513. PMID 32403331.
- ^ Soto SM (April 2013). "Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm". Virulence. 4 (3): 223–9. doi:10.4161/viru.23724. PMC 3711980. PMID 23380871.
- ^ a b Cai Y, Wang C, Chen Z, Xu Z, Li H, Li W, et al. (August 2020). "Transporters HP0939, HP0497, and HP0471 participate in intrinsic multidrug resistance and biofilm formation in Helicobacter pylori by enhancing drug efflux". Helicobacter. 25 (4): e12715. doi:10.1111/hel.12715. PMID 32548895. S2CID 219726485. Archived from the original on 15 February 2024. Retrieved 15 February 2024.
- ^ Mommersteeg MC, Nieuwenburg SA, Wolters LM, Roovers BH, van Vuuren HA, Verhaar AP, et al. (November 2023). "The use of non-invasive stool tests for verification of Helicobacter pylori eradication and clarithromycin resistance". United European Gastroenterol J. 11 (9): e894-903. doi:10.1002/ueg2.12473. PMC 10637120. PMID 37854002.
- ^ Stenström B, Mendis A, Marshall B (August 2008). "Helicobacter pylori--the latest in diagnosis and treatment". Australian Family Physician. 37 (8): 608–12. PMID 18704207.
- ^ Fischbach L, Evans EL (August 2007). "Meta-analysis: the effect of antibiotic resistance status on the efficacy of triple and quadruple first-line therapies for Helicobacter pylori". Alimentary Pharmacology & Therapeutics (Meta-analysis). 26 (3): 343–57. doi:10.1111/j.1365-2036.2007.03386.x. PMID 17635369. S2CID 20973127.
- ^ Graham DY, Shiotani A (June 2008). "New concepts of resistance in the treatment of Helicobacter pylori infections". Nature Clinical Practice. Gastroenterology & Hepatology. 5 (6): 321–31. doi:10.1038/ncpgasthep1138. PMC 2841357. PMID 18446147.
- ^ Pohl D, Keller PM, Bordier V, Wagner K (August 2019). "Review of current diagnostic methods and advances in Helicobacter pylori diagnostics in the era of next generation sequencing". World J Gastroenterol. 25 (32): 4629–4660. doi:10.3748/wjg.v25.i32.4629. PMC 6718044. PMID 31528091.
- ^ Sukri A, Hanafiah A, Patil S, Lopes BS (April 2023). "The Potential of Alternative Therapies and Vaccine Candidates against Helicobacter pylori". Pharmaceuticals. 16 (4): 552. doi:10.3390/ph16040552. PMC 10141204. PMID 37111309.
- ^ a b c Vaou N, Stavropoulou E, Voidarou C, Tsigalou C, Bezirtzoglou E (September 2021). "Towards Advances in Medicinal Plant Antimicrobial Activity: A Review Study on Challenges and Future Perspectives". Microorganisms. 9 (10): 2041. doi:10.3390/microorganisms9102041. PMC 8541629. PMID 34683362.
- ^ Moon JK, Kim JR, Ahn YJ, Shibamoto T (June 2010). "Analysis and anti-Helicobacter activity of sulforaphane and related compounds present in broccoli ( Brassica oleracea L.) sprouts". Journal of Agricultural and Food Chemistry. 58 (11): 6672–7. doi:10.1021/jf1003573. PMID 20459098.
- ^ Sathianarayanan S, Ammanath AV, Biswas R, B A, Sukumaran S, Venkidasamy B (July 2022). "A new approach against Helicobacter pylori using plants and its constituents: A review study". Microb Pathog. 168: 105594. doi:10.1016/j.micpath.2022.105594. PMID 35605740. S2CID 248975163.
- ^ Moradi Y, Majidi L, Khateri S, Azh N, Gheshlagh RG, Saniee N, et al. (July 2023). "The association between periodontal diseases and helicobacter pylori: an updated meta-analysis of observational studies". BMC Oral Health. 23 (1): 523. doi:10.1186/s12903-023-03232-3. PMC 10369707. PMID 37496045.
- ^ Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F (August 2020). "Gastric cancer". Lancet. 396 (10251): 635–648. doi:10.1016/S0140-6736(20)31288-5. PMID 32861308.
- ^ Badgwell B, Das P, Ajani J (August 2017). "Treatment of localized gastric and gastroesophageal adenocarcinoma: the role of accurate staging and preoperative therapy". Journal of Hematology & Oncology. 10 (1): 149. doi:10.1186/s13045-017-0517-9. PMC 5558742. PMID 28810883.
- ^ Laird-Fick HS, Saini S, Hillard JR (August 2016). "Gastric adenocarcinoma: the role of Helicobacter pylori in pathogenesis and prevention efforts". Postgraduate Medical Journal. 92 (1090): 471–7. doi:10.1136/postgradmedj-2016-133997. PMID 27222587. S2CID 20739020.
- ^ Bron D, Meuleman N (September 2019). "Marginal zone lymphomas: second most common lymphomas in older patients". Current Opinion in Oncology. 31 (5): 386–393. doi:10.1097/CCO.0000000000000554. PMID 31246587. S2CID 195765608.
- ^ Kobayashi T, Takahashi N, Hagiwara Y, Tamaru J, Kayano H, Jin-nai I, et al. (January 2008). "Successful radiotherapy in a patient with primary rectal mucosa-associated lymphoid tissue lymphoma without the API2-MALT1 fusion gene: a case report and review of the literature". Leukemia Research. 32 (1): 173–5. doi:10.1016/j.leukres.2007.04.017. PMID 17570523.
- ^ Matysiak-Budnik T, Priadko K, Bossard C, Chapelle N, Ruskoné-Fourmestraux A (July 2023). "Clinical Management of Patients with Gastric MALT Lymphoma: A Gastroenterologist's Point of View". Cancers. 15 (15): 3811. doi:10.3390/cancers15153811. PMC 10417821. PMID 37568627.
- ^ Casulo C, Friedberg J (2017). "Transformation of marginal zone lymphoma (and association with other lymphomas)". Best Practice & Research. Clinical Haematology. 30 (1–2): 131–138. doi:10.1016/j.beha.2016.08.029. PMID 28288708.
- ^ a b c Kuo SH, Yeh KH, Chen LT, Lin CW, Hsu PN, Hsu C, et al. (June 2014). "Helicobacter pylori-related diffuse large B-cell lymphoma of the stomach: a distinct entity with lower aggressiveness and higher chemosensitivity". Blood Cancer Journal. 4 (6): e220. doi:10.1038/bcj.2014.40. PMC 4080211. PMID 24949857.
- ^ a b c Cheng Y, Xiao Y, Zhou R, Liao Y, Zhou J, Ma X (August 2019). "Prognostic significance of helicobacter pylori-infection in gastric diffuse large B-cell lymphoma". BMC Cancer. 19 (1): 842. doi:10.1186/s12885-019-6067-5. PMC 6712724. PMID 31455250.
- ^ Tsai HJ, Tai JJ, Chen LT, Wu MS, Yeh KH, Lin CW, et al. (July 2020). "A multicenter prospective study of first-line antibiotic therapy for early-stage gastric mucosa-associated lymphoid tissue lymphoma and diffuse large B-cell lymphoma with histological evidence of mucosa-associated lymphoid tissue". Haematologica. 105 (7): e349–e354. doi:10.3324/haematol.2019.228775. PMC 7327622. PMID 31727764.
- ^ Balendra V, Amoroso C, Galassi B, Esposto J, Bareggi C, Luu J, et al. (August 2023). "High-Salt Diet Exacerbates H. pylori Infection and Increases Gastric Cancer Risks". J Pers Med. 13 (9): 1325. doi:10.3390/jpm13091325. PMC 10533117. PMID 37763093.
- ^ Jaroenlapnopparat A, Bhatia K, Coban S (June 2022). "Inflammation and Gastric Cancer". Diseases. 10 (3): 35. doi:10.3390/diseases10030035. PMC 9326573. PMID 35892729.
- ^ Park JM, Han YM, Oh JY, Lee DY, Choi SH, Hahm KB (September 2021). "Transcriptome profiling implicated in beneficiary actions of kimchi extracts against Helicobacter pylori infection". J Clin Biochem Nutr. 69 (2): 171–187. doi:10.3164/jcbn.20-116. PMC 8482382. PMID 34616109.
- ^ a b Brown LM (2000). "Helicobacter pylori: epidemiology and routes of transmission". Epidemiologic Reviews. 22 (2): 283–97. doi:10.1093/oxfordjournals.epirev.a018040. PMID 11218379.
- ^ Pacifico L, Osborn JF, Bonci E, Romaggioli S, Baldini R, Chiesa C (January 2014). "Probiotics for the treatment of Helicobacter pylori infection in children". World J Gastroenterol. 20 (3): 673–83. doi:10.3748/wjg.v20.i3.673. PMC 3921477. PMID 24574741.
- ^ Goodman KJ, O'rourke K, Day RS, Wang C, Nurgalieva Z, Phillips CV, et al. (December 2005). "Dynamics of Helicobacter pylori infection in a US-Mexico cohort during the first two years of life". International Journal of Epidemiology. 34 (6): 1348–55. doi:10.1093/ije/dyi152. PMID 16076858.
- ^ Li R, Zhang P, Hu Z, Yi Y, Chen L, Zhang H (14 May 2021). "Helicobacter pylori reinfection and its risk factors after initial eradication: A protocol for systematic review and meta-analysis". Medicine. 100 (19): e25949. doi:10.1097/MD.0000000000025949. PMC 8133036. PMID 34106668.
- ^ a b c Blaser MJ (February 2005). "An endangered species in the stomach". Scientific American. 292 (2): 38–45. Bibcode:2005SciAm.292b..38B. doi:10.1038/scientificamerican0205-38. PMID 15715390.
- ^ Graham DY, Yamaoka Y, Malaty HM (November 2007). "Contemplating the future without Helicobacter pylori and the dire consequences hypothesis". Helicobacter. 12 (Suppl 2): 64–8. doi:10.1111/j.1523-5378.2007.00566.x. PMC 3128250. PMID 17991179.
- ^ Delaney B, McColl K (August 2005). "Review article: Helicobacter pylori and gastro-oesophageal reflux disease". Alimentary Pharmacology & Therapeutics (Review). 22 (Suppl 1): 32–40. doi:10.1111/j.1365-2036.2005.02607.x. PMID 16042657. S2CID 34921548.
- ^ Blaser MJ, Chen Y, Reibman J (May 2008). "Does Helicobacter pylori protect against asthma and allergy?". Gut. 57 (5): 561–7. doi:10.1136/gut.2007.133462. PMC 3888205. PMID 18194986.
- ^ Chen Y, Blaser MJ (August 2008). "Helicobacter pylori colonization is inversely associated with childhood asthma". The Journal of Infectious Diseases. 198 (4): 553–60. doi:10.1086/590158. PMC 3902975. PMID 18598192.
- ^ Smoak BL, Kelley PW, Taylor DN (March 1994). "Seroprevalence of Helicobacter pylori infections in a cohort of US Army recruits". American Journal of Epidemiology. 139 (5): 513–9. doi:10.1093/oxfordjournals.aje.a117034. PMID 8154475.
- ^ Everhart JE, Kruszon-Moran D, Perez-Perez GI, Tralka TS, McQuillan G (April 2000). "Seroprevalence and ethnic differences in Helicobacter pylori infection among adults in the United States". The Journal of Infectious Diseases. 181 (4): 1359–63. doi:10.1086/315384. PMID 10762567.
- ^ Malaty HM (2007). "Epidemiology of Helicobacter pylori infection". Best Practice & Research. Clinical Gastroenterology. 21 (2): 205–14. doi:10.1016/j.bpg.2006.10.005. PMID 17382273.
- ^ Mégraud F (September 2004). "H pylori antibiotic resistance: prevalence, importance, and advances in testing". Gut. 53 (9): 1374–84. doi:10.1136/gut.2003.022111. PMC 1774187. PMID 15306603.
- ^ Correa P, Piazuelo MB (January 2012). "Evolutionary History of the Helicobacter pylori Genome: Implications for Gastric Carcinogenesis". Gut and Liver. 6 (1): 21–8. doi:10.5009/gnl.2012.6.1.21. PMC 3286735. PMID 22375167.
- ^ Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, et al. (February 2007). "An African origin for the intimate association between humans and Helicobacter pylori". Nature. 445 (7130): 915–918. Bibcode:2007Natur.445..915L. doi:10.1038/nature05562. PMC 1847463. PMID 17287725.
- ^ "The Nobel Prize in Physiology or Medicine 2005". Archived from the original on 23 May 2020. Retrieved 30 August 2018.
- ^ Bizzozero G (1893). "Ueber die schlauchförmigen Drüsen des Magendarmkanals und die Beziehungen ihres Epitheles zu dem Oberflächenepithel der Schleimhaut". Archiv für Mikroskopische Anatomie. 42: 82–152. doi:10.1007/BF02975307. S2CID 85338121. Archived from the original on 2 December 2020. Retrieved 29 June 2019.
- ^ Konturek JW (December 2003). "Discovery by Jaworski of Helicobacter pylori and its pathogenetic role in peptic ulcer, gastritis and gastric cancer" (PDF). Journal of Physiology and Pharmacology. 54 (Suppl 3): 23–41. PMID 15075463. Archived from the original (PDF) on 30 September 2004. Retrieved 25 August 2008.
- ^ Egan BJ, O'Morain CA (2007). "A historical perspective of Helicobacter gastroduodenitis and its complications". Best Practice & Research. Clinical Gastroenterology. 21 (2): 335–46. doi:10.1016/j.bpg.2006.12.002. PMID 17382281.
- ^ Palmer ED (August 1954). "Investigation of the gastric mucosa spirochetes of the human". Gastroenterology. 27 (2): 218–20. doi:10.1016/S0016-5085(19)36173-6. PMID 13183283.
- ^ Steer HW (August 1975). "Ultrastructure of cell migration throught [sic] the gastric epithelium and its relationship to bacteria". Journal of Clinical Pathology. 28 (8): 639–46. doi:10.1136/jcp.28.8.639. PMC 475793. PMID 1184762.
- ^ Marshall BJ, Warren JR (June 1984). "Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration". Lancet. 1 (8390): 1311–5. doi:10.1016/S0140-6736(84)91816-6. PMID 6145023. S2CID 10066001.
- ^ Atwood KI (2004). "Bacteria, Ulcers, and Ostracism? H. pylori and the making of a myth". Archived from the original on 5 November 2009. Retrieved 2 August 2008.
- ^ Helicobacter pylori in peptic ulcer disease (Report). NIH Consensus Statement Online. Vol. 12. 7–9 January 1994. pp. 1–23. Archived from the original on 19 August 2011. Retrieved 21 December 2004.
- ^ a b Liddell HG, Scott R (1966). A Lexicon: Abridged from Liddell and Scott's Greek-English Lexicon. Oxford, UK: Oxford University Press. ISBN 978-0-19-910207-5.
- ^ Marshall BS, Goodwin CS (1987). "Revised nomenclature of Campylobacter pyloridis". International Journal of Systematic Bacteriology. 37 (1): 68. doi:10.1099/00207713-37-1-68.
- ^ Goodwin CS, Armstrong JA, Chilvers T, Peters M, Collins MD, Sly L, et al. (1989). "Transfer of Campylobacter pylori and Campylobacter mustelae to Helicobacter gen. nov. as Helicobacter pylori comb. nov. and Helicobacter mustelae comb. nov. respectively". International Journal of Systematic Bacteriology. 39 (4): 397–405. doi:10.1099/00207713-39-4-397.
- ^ Buckley MJ, O'Morain CA (1998). "Helicobacter biology--discovery". British Medical Bulletin. 54 (1): 7–16. doi:10.1093/oxfordjournals.bmb.a011681. PMID 9604426.
- ^ Mégraud F, et al. (European Helicobacter Study Group) (November 2007). "Evolution of Helicobacter pylori research as observed through the workshops of the European Helicobacter Study Group". Helicobacter. 12 Suppl 2 (Suppl 2): 1–5. doi:10.1111/j.1523-5378.2007.00581.x. PMID 17991169. S2CID 45841196.
- ^ "EHMSG". Ehmsg2019. Archived from the original on 11 January 2024. Retrieved 11 January 2024.
- ^ Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, et al. (European Helicobacter Study Group) (May 2012). "Management of Helicobacter pylori infection--the Maastricht IV/ Florence Consensus Report". Gut. 61 (5): 646–64. doi:10.1136/gutjnl-2012-302084. hdl:1765/64813. PMID 22491499. S2CID 1401974. Archived from the original on 4 July 2021. Retrieved 7 December 2012.
- ^ Malfertheiner P, Megraud F, O'Morain C, Bazzoli F, El-Omar E, Graham D, et al. (June 2007). "Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report". Gut. 56 (6): 772–81. doi:10.1136/gut.2006.101634. PMC 1954853. PMID 17170018.
- ^ Malfertheiner P, Mégraud F, O'Morain C, Hungin AP, Jones R, Axon A, et al. (European Helicobacter Pylori Study Group (EHPSG)) (February 2002). "Current concepts in the management of Helicobacter pylori infection--the Maastricht 2-2000 Consensus Report". Alimentary Pharmacology & Therapeutics. 16 (2): 167–80. doi:10.1046/j.1365-2036.2002.01169.x. PMID 11860399. S2CID 6166458.
- ^ Malfertheiner P, Mégraud F, O'Morain C, Bell D, Bianchi Porro G, Deltenre M, et al. (European Helicobacter Pylori Study Group (EHPSG)) (January 1997). "Current European concepts in the management of Helicobacter pylori infection--the Maastricht Consensus Report. The European Helicobacter Pylori Study Group (EHPSG)". European Journal of Gastroenterology & Hepatology. 9 (1): 1–2. doi:10.1097/00042737-199701000-00002. PMID 9031888. S2CID 36930542.
- ^ McNicholl AG, Gasbarrini A, Tepes B, et al. (September 2014). "Pan-European registry on H. pylori management (Hp-EuReg): Interim analysis of 5,792 patients". Helicobacter. 2014: 69.
- ^ "Management of Helicobacter pylori infection". Online courses. United European Gastroenterology. Archived from the original on 2 April 2015.
- ^ "Annual Report 2012". United European Gastroenterology. Archived from the original on 4 June 2016. Retrieved 25 February 2015.
- ^ Jung SW, Lee SW (January 2016). "The antibacterial effect of fatty acids on Helicobacter pylori infection". The Korean Journal of Internal Medicine (Review). 31 (1): 30–5. doi:10.3904/kjim.2016.31.1.30. PMC 4712431. PMID 26767854.
External links
[edit]- "Information on tests for H. pylori". National Institutes of Health. U.S. Department of Health and Human Services. Archived from the original on 13 June 2013.
- "European Helicobacter Study Group (EHSG)".
- "Type strain of Helicobacter pylori at BacDive". Bacterial Diversity Metadatabase.
- "Helicobacter pylori". Genome. KEGG. Japan. 26695.
