Talk:Neutral mutation
| This article is rated C-class on Wikipedia's content assessment scale. It is of interest to the following WikiProjects: | |||||||||||||||||
| |||||||||||||||||
Minor ?
[edit]"A neutral mutation may or may not affect the resulting protein, but will do so in a very minor fashion". I removed the part in bold, it's simply not true. A mutation could have a huge impact on an organism (destroying an organ), and it could still be neutral (if the organ is useless at the moment). - PhDP (talk) 22:51, 11 July 2007 (UTC)
Mutation Table
[edit]I'm not sure if that table titled mutation helps the article at all. Is there a place for it here? Grabriggs (talk) 00:08, 22 October 2013 (UTC)
Suggested Improvements
[edit]1) I am working with a partner to improve this article for a class. We are thinking about adding sections titled history, synonomous mutations, measurements of neutrality and impact on evolutionary theory. We will then expound on these topics. Grabriggs (talk) 00:25, 22 October 2013 (UTC)
2) The lead is not sourced and should be updated with references. If no one has issue with this I will delete the lead and start over or substantially change it and provide references. Grabriggs (talk) 05:50, 25 October 2013 (UTC)
Proposed New Lead
[edit]Mutations in which natural selection is not influential regarding fixation are termed neutral mutations. (1) Evolution is driven by mutations to the genome of organisms. Some mutations may have a negative or positive impact on the organism which in turn will effect the ability to survive in the environment. The process of natural selection is responsible for the fixation or deletion of mutated genes over time. A gene that has a positive impact on the organism is more likely to replace the original version of the gene in subsequent generations and become fixed in the species‘ genome. Genes in which mutations cause a negative effect will be more likely to be deleted from the gene pool in future generations. A mutation that gives no advantage or disadvantage to the organism is not pushed by natural selection and is termed neutral. Neutral mutations are random in their ability to become fixed in the genomes of species. (2)
1) Tomizawa J. (2000) Derivation of the relationship between neutral mutation and fixation solely from the definition of selective neutrality. PNAS June 20, 2000 vol. 97 no. 13 7372-7375.
2) Duret, L. (2008) Neutral theory: The null hypothesis of molecular evolution. Nature Education 1(1)
Grabriggs (talk) 01:51, 26 October 2013 (UTC)
- The proposed new lede is not focused enough on the topic of the article, neutral mutations. Over half of the proposed lede discusses mutations under selection. Joannamasel (talk) 01:55, 26 October 2013 (UTC)
- I've reworked and simplified what my partner started with. We will be adding more as the project continues. Any feedback is appreciated.
Neutral mutations are changes in DNA sequence that are not beneficial nor detrimental to the fitness of an organism.
In population genetics, mutations in which natural selection is not influential regarding fixation are termed neutral mutations. (1) If inheritable, a neutral mutation at an unlinked locus will proceed to loss or fixation in the genome based on random sampling known as genetic drift. (2)
Mutations that do not influence the active site of a protein can also be characterized as neutral mutations. (3)
The majority of substitutions in a genome appear to have no visible effect on the fitness of individuals and are therefore neutral. (4,5)
1. Tomizawa J. (2000) Derivation of the relationship between neutral mutation and fixation solely from the definition of selective neutrality. PNAS June 20, 2000 vol. 97 no. 13 7372-7375.
2. Nei, M., Suzuki, Y., & Nozawa, M. (2010). The neutral theory of molecular evolution in the genomic era. Annual Review of Genomics & Human Genetics, 11(1), 265.
3. Kimura, M. (1968). Evolutionary rate at the molecular level. Nature, 217(5129), 624.
4. Gillespie, J. H. (1991). The causes of molecular evolution. New York: Oxford University Press.
5. Kimura, M. (1983). The neutral theory of molecular evolution. Cambridge Cambridgeshire ;New York: Cambridge University Press.
Seanmcaruthers (talk) 04:00, 28 October 2013 (UTC)
Proposed Additions
[edit]Lead
Mutations in which natural selection is not influential regarding fixation are termed neutral mutations. (1) The process of natural selection is responsible for the fixation or deletion of mutated genes over time. A gene that has a positive impact on the organism is more likely to replace the original version of the gene in subsequent generations and become fixed in the species‘ genome. Genes in which mutations cause a negative effect will be more likely to be deleted from the gene pool in future generations. A third possibility also exists. A mutation that gives no advantage or disadvantage to the organism is termed a neutral mutation. Since neutral mutations are not pushed by natural selection they are random in their ability to become fixed in the genomes of species. The random change in allele frequencies from generation to generation is referred to as genetic drift. A neutral mutation and its potential fixation are tied to the genetic drift in an organism’s genome. The neutral theory of molecular evolution considers neutral mutation and random genetic drift over time in organisms. (2)
History
"Variations neither useful nor injurious would not be affected by natural selection, and would be left either a fluctuating element, as perhaps we see in certain polymorphic species, or would ultimately become fixed, owing to the nature of the organism and the nature of the conditions." Darwin commented on the idea of neutral mutation in his work hypothesizing that mutations that do not give an advantage or disadvantage may fluctuate or become fixed apart from natural selection. (3)
Synonymous Mutation
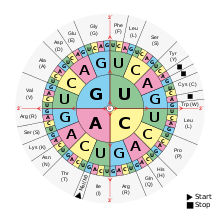
When an incorrect base is inserted during replication or transcription of a coding region it can effect the eventual translation of the sequence into amino acids. However, since multiple codons are used for the same amino acids a change in a single base may still lead to translation of the same amino acid. The changing of a base in a codon without the changing of the translated amino acid is called a synonymous mutation. (5) Some research has suggested that there is bias in selection of base substitution in synonymous mutation. This could be due to selective pressure to improve translation efficiency associated with the most available tRNAs or simply mutational bias. (6) If these mutations influence the rate of translation or an organism’s ability to manufacture protein these synonymous mutations may actually influence the fitness of the effected organism. (5)
Impact on Evolutionary Theory
Neutral mutation has become a part of the neutral theory of molecular evolution, proposed in the 1960s. This theory suggests that neutral mutations are responsible for a large portion of an organism’s evolution and phenotype. For example, bovine and human insulin, while differing in amino acid sequence are still able to perform the same function. The amino acid substitutions between species were seen therefore to be neutral or not impactful to the function of the protein. (4) A number of impacts due to neutral mutation were predicted in neutral theory including: amino acids with similar biochemical properties should be substituted more often than biochemically different amino acids; synonymous base substitutions should be observed more often than nonsynonymous substitutions; introns should evolve at the same rate as synonymous mutations in coding exons; and pseudogenes should also evolve at a similar rate. These predictions have been confirmed with the introduction of additional genetic data since the theory’s introduction. (2)
References to be inserted inline
1) Tomizawa J. (2000) Derivation of the relationship between neutral mutation and fixation solely from the definition of selective neutrality. PNAS June 20, 2000 vol. 97 no. 13 7372-7375.
2) Duret, L. (2008) Neutral theory: The null hypothesis of molecular evolution. Nature Education 1(1)
3) Darwin, C. (1987; 1859). On the origin of species by means of natural selection : Or the preservation of favoured races in the struggle for life (Special ed.). Birmingham, Ala.: Gryphon Editions.
4)Nei, M., Suzuki, Y., & Nozawa, M. (2010). The neutral theory of molecular evolution in the genomic era. Annual Review of Genomics & Human Genetics, 11(1), 265.
5) Venetianer, P. á. (2012). Are synonymous codons indeed synonymous? Biomolecular Concepts, 3(1), 21.
6) Duret, L. (2002) Evolution of synonymous codon usage in metazoans. Current Opinion in Genetics & Development. Volume 12, Issue 6, 1 December 2002, Pages 640–649.
Grabriggs (talk) 03:44, 28 October 2013 (UTC)
- Measurement of neutrality
Neutral mutations are measured in population and evolutionary genetics often by looking at variation in populations. These have been measured historically by gel electrophoresis to determine allozyme frequencies (1). Statistical analyses of this data is used to compare variation to predicted values based on population size, mutation rates and effective population size. The accumulation of data based on observed polymorphism led to the formation of the neutral theory of evolution. (2) According to the neutral (often the nearly neutral) theory of evolution, the rate of fixation in a population of a neutral mutation will be directly related to the rate of formation of the neutral allele. The McDonald-Kreitman test (3) has been used to study selection over long periods of evolutionary time. The test compares variation within a species to variation among species by comparing the rate of variation in a neutral site to that of a non-neutral site. The test often uses synonymous substitutions in protein coding genes as the neutral component, however, synonymous mutations been shown to be under purifying selection in many instances. (4, 5)
Selection studies
Mutational experiments in laboratory settings using Caenorhabditis elegans with deficient DNA repair mechanisms have shown that much less than 1% of mutations show on effect on fitness and thus could be called neutral mutations. (6) Site directed mutagenesis and mutation accumulation studies in RNA viruses indicate that 55-60% of observable mutations could be considered neutral, noting that the high gene density of RNA viruses makes them among the least tolerant of mutational stress of all life. (7, 8)
1. Lewontin, R. C. (1991). Twenty-five years ago in genetics: Electrophoresis in the development of evolutionary genetics: Milestone or millstone? Genetics, 128(4), 657.
2. Kimura, M. (1968). Evolutionary rate at the molecular level. Nature, 217(5129), 624.
3. McDonald, J. H., & Kreitman, M. (1991). Adaptive protein evolution at the adh locus in drosophila. Nature, 351(6328), 652.
4. Hellmann, I., Zollner, S., Enard, W., Ebersberger, I., Nickel, B., & Paabo, S. (2003). Selection on human genes as revealed by comparisons to chimpanzee cDNA. Genome Research, 13(5), 831.
5. Zhou, T., Gu, W., & Wilke, C. O. (2010). Detecting positive and purifying selection at synonymous sites in yeast and worm. Molecular Biology and Evolution, 27(8), 1912.
6. Estes, S., Phillips, P. C., Denver, D. R., Thomas, W. K., & Lynch, M. (2004). Mutation accumulation in populations of varying size: The distribution of mutational effects for fitness correlates in caenorhabditis elegans. Genetics, 166(3), 1269.
7. Burch, C. L., Guyader, S., & Samarov, D. (2007). Experimental estimate of the abundance and effects of nearly neutral mutations in the RNA virus F6. Genetics, 176(1), 467.
8. Sanjuán, R., Moya, A., & Elena, S. F. (2004). The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proceedings of the National Academy of Sciences of the United States of America, 101(22), 8396.
Seanmcaruthers (talk) 08:00, 28 October 2013 (UTC)
Comments from Klortho
[edit]- General:
- Very nice, overall! You have added a good amount of content, as well as a figure, and you have a lot of links and references.
- Headings should follow WP style: only the first word should be capitalized.
- You could add a few more intra-wiki links (but don't overdo it). For example, "amino acid" doesn't seem to be linked anywhere.
- Lead:
- In general, written at a bit too technical a level. For example, "loss" or "fixation" -- I think most people who are not biologists probably don't know what these mean. Ditto for "allele". You should be able to find simpler language for some of these concepts, and/or gloss these terms as they're introduced.
- Why is this concept notable? What makes neutral mutations important?
- History:
- I would flip the paragraph around so that you mention Darwin first, then make the quotation into an indented block-quote.
- Use his full name, Charles Darwin.
- Any more history, or does it end with Darwin?
- Synonymous mutation
- This seems to be a rather abrupt change in topic. It throws the reader off -- what are you talking about now?
- Should it be a subsection of a larger section, "Types of neutral mutations?" Are there other types? You should probably give each major type its own subsection.
- You never say that anyone thinks that synonymous mutations *are* neutral mutations. At the very end, you hint that maybe they are not, but after reading the paragraph, I'm left not knowing how these are related to the topic of neutral mutations.
- Impact on Evolutionary Theory
- You mention the "neutral theory of molecular evolution", but don't give any indication of why it is notable -- i.e., how is it different from what anyone else thinks/thought?
- Could you elaborate a bit on the last sentence: how were the predictions confirmed?
- Selection studies
- I'm confused by this: "much less than 1% of mutations show on effect on fitness and thus could be called neutral mutations". If < 1% of mutations show an effect on fitness, doesn't that mean that >99% of mutations are neutral?
- The next paragraph describes 55-60% are neutral. Why the discrepancy? Rather than just throw out facts, you should try to have a narrative, and use the facts to explain things.
- References: Please use the procedure described here for formatting your references. First look them up in PubMed, and then use the magnifying glass tool to fill in the form.
Klortho (talk) 06:36, 3 November 2013 (UTC)
- Thanks for the comments Klortho! We will be working to improve the article this week. I've started updating the history section to add some depth to the subject and will be addressing a number of other suggestions you added. Grabriggs (talk) 00:17, 6 November 2013 (UTC)
- I also added a section on types of neutral mutations with two subsections underneath describing two different types along with a picture of Darwin Grabriggs (talk) 04:14, 6 November 2013 (UTC)
- I like the suggestions on the impact to evolutionary theory. I'm going to work on updating that section this week. Grabriggs (talk) 01:37, 12 November 2013 (UTC)
- Thanks for the review and advice. I tried to clean up the intro a little, but still might be some technical jargon. I got rid of the selection studies part as it didn't fit in well and seemed to confuse everyone.Seanmcaruthers (talk) 03:56, 12 November 2013 (UTC)
- I like the suggestions on the impact to evolutionary theory. I'm going to work on updating that section this week. Grabriggs (talk) 01:37, 12 November 2013 (UTC)
- I also added a section on types of neutral mutations with two subsections underneath describing two different types along with a picture of Darwin Grabriggs (talk) 04:14, 6 November 2013 (UTC)
Comments from Jfitz1974
[edit]The changes you have made are improvements far and above the prior version of the page. The amount of material added and the addition of a graphic greatly add to the substance of the article. Here are some suggestions I have.
Lead Section
In the opening sentence of the lead, rather than saying “not beneficial nor detrimenta,” I think that using the neither / nor combination makes the statement a bit easier to follow.
The last statement discussing the “majority of substitutions” might raise some questions of what is being substituted. Looking at it from the point of view from a non-science person, just saying substitution could lead them to think of something other than a base-substitution mutation.
History Section
Opening with a direct quote seems out of place to me. The first thing that ran through my mind when I began reading the section was: who is saying this? I knew that it was a direct quote because of the use of quotation marks, but I was questioning if the person who said it was someone who knew what they were talking about with regard to mutation and molecular biology. Starting off by saying that it is a direct quote from Charles Darwin would have really grabbed my attention and made me appreciate the quote more.
Synonymous Mutation Section
I find the last sentence to contain some redundancy in the wording. You start by saying “these mutations” and then later in the same sentence state “these synonymous mutations.” Perhaps to ease the redundancy of “these” you could start by simply using “synonymous mutations” and then later use “these mutations.” Also, I believe the appropriate term to use is “affected organisms” over “effected organisms” in this situation.
I like the graphic that is used in this section, but would a table illustrating the different amino acids and all the codons that code for each would drive home the point that many amino acids are coded for by more than one codon? For example, Serine would be in one field of the table along with all of the codons that code for it.
Impact on Evolutionary Theory Section
I really like the insulin example as an example of a neutral mutation, however in your leader you define that a neutral mutation doesn’t affect the fitness of an organism (which usually refers to an individual’s reproductive success). I found a biology textbook (Biology: Core Concepts, 2nd edition by Levine and Miller) that define a neutral mutations as “mutations that do not change the functional characteristic of the protein that the genes specify.” The use of the insulin example would fit in perfectly with this type of definition.
Measurement of Neutrality Section
After reading this section, I am still not quite sure exactly what a measurement of neutrality is. Is neutrality the rate of occurrence of neutral mutations in a population or organism?
Is the neutral theory that is linked in this section the same as the neutral theory of molecular evolution? The link to the Wiki page neutral theory wasn’t very clear on that.
Selection Studies Section
What is the purpose of the selection studies section? What do you want the reader to know from this section? After reading the Estes article that is cited, I got the impression that the authors were looking at relationship between population size and mutation accumulation- specifically as they pertain to the decline in fitness. They discussed mutations that created large effects organisms and mutations that cause small effects in organisms, but didn’t mention the mutations that produce no effects in organisms. I’m not sure this is the best study to use when discussing neutral mutations.
Other Suggestions
Based upon Klortho’s suggestion for more intra-Wiki links, here are some items that could possibly be used.
- Fitness
- Locus
- Genome
- Allele
- Amino Acid
- Intron
- Exon
- Pseudogenes
- Polymorphism — Preceding unsigned comment added by Jfitz1974 (talk • contribs) 03:24, 4 November 2013 (UTC)
- Thanks for the suggestions! A number of comments mentioned the figure that was inserted so I have substituted a more simple table instead that shows the different codons for each amino acid Grabriggs (talk) 04:18, 6 November 2013 (UTC)
- Thanks. I tried to address most of the comments. The selection studies section is gone and I hopefully added to the measurement section that clarifies.Seanmcaruthers (talk) 04:04, 12 November 2013 (UTC)
- Great job guys! I really like the changes you've made, and think the new genetic code graphic is wonderful!! Jfitz1974 (talk) 19:17, 11 December 2013 (UTC)
- Thanks. I tried to address most of the comments. The selection studies section is gone and I hopefully added to the measurement section that clarifies.Seanmcaruthers (talk) 04:04, 12 November 2013 (UTC)
- Thanks for the suggestions! A number of comments mentioned the figure that was inserted so I have substituted a more simple table instead that shows the different codons for each amino acid Grabriggs (talk) 04:18, 6 November 2013 (UTC)
Comments from Rebecca Chappel
[edit]You have some great comments on here already - I'll try to add some useful ones.
- The organization of the article looks great. I like the quote from Darwin in the history section. The history section is a nice addition I would not have thought to add.
- For the Synonymous Mutation Section: I like the image but if I were new to this subject the figure might be confusing. I think a nice addition would be a simple chart of all of the amino acids and their respective bases to see how it is possible for a base to change without changing the amino acid (in addition to the current image).
- The addition of a polymorphism link would be nice in case the reader needs to reference the subject. (on the Measurement of Neutrality section). Overall, I think the links you chose are good.
- I really like the addition of a practical example – the bovine and human insulin example is great.
- Overall – some questions I would have if this were the only knowledge I had on the subject:
- Are there Neutral Mutations that are not Synonymous Mutations? Are there any other kinds?
- The Selection studies section is a little confusing – is there any additional information about the studies?
- I think it looks great! A huge improvement compared to the information that was on there before. Rebeccachappel (talk) 21:03, 5 November 2013 (UTC)
- Thanks for the comments. I've added some additional information related to synonymous and neutral mutations. I think you had a great idea that that section needed more explanation! Grabriggs (talk) 00:14, 9 November 2013 (UTC)
- Thanks for the helpful insight. You had a lot of the same concerns as the others and hopefully we are beginning to address them all.Seanmcaruthers (talk) 04:04, 12 November 2013 (UTC)
- Thanks for the comments. I've added some additional information related to synonymous and neutral mutations. I think you had a great idea that that section needed more explanation! Grabriggs (talk) 00:14, 9 November 2013 (UTC)
Comments from Harshil Patel
[edit]I see there adequate progress on the article, good work on that. I noticed that the headline for 'Selection studies' has a different format. It would be wise to have the same format of the heading throughout you article. I like how you have a history section for this article since the content is theory based. You have put in work to this article with pictures and examples and adequate amount of reference used. Keep going. Igenes (talk) 01:09, 6 November 2013 (UTC)
- Thanks for the comments.Seanmcaruthers (talk) 04:04, 12 November 2013 (UTC)
As we move forward through this semester, I have additional comments to improve your article page. More to come as we move along.
- I think adding a section on 'Application' of the neutral mutation would help the article page in gaining more content.
- Include some examples of Neutral mutation along with some pictures - You may want to have it under each 'Types of netural mutation'.
- In you "History" section you mentioned the work of Motoo Kimura - I suggest there should be a wikilink to his work.
Igenes (talk) 19:24, 22 November 2013 (UTC)
- Thanks again. I have added substantially to the Molecular clocks section which is a major application of neutral mutations. Are there other applications you were considering that we are missing, or just think we could expand on what we have if we put it in a new section?Seanmcaruthers (talk) 07:11, 26 November 2013 (UTC)
What I meant by applications of neutral mutations is that giving few examples where neutral mutation is seen. Such as vestigial organs are linked to neutral mutation - one such example is 'Six toes'. I think this would help the article to gain more content and interest.Igenes (talk) 16:07, 27 November 2013 (UTC)
- Under 'Synonymous mutation of bases' - you mention about wobble concept. I think there should be a wikilink to the wooble concept.
- Under 'Synonymous mutation of bases' - I really like your color worded codon table, however I think there should be a small description and name for the table.Igenes (talk) 01:56, 5 December 2013 (UTC)
Neutral theory
[edit]Great job guys. One thing that needs work is that neutral theory does not claim that the majority of mutations are neutral. Instead, it claims only that the majority of the mutations that we can detect as divergence or polymorphism are neutral. It allows for large numbers of deleterious mutations, but assumes that these are removed so rapidly by natural selection that we don't see them. This contradicts a few statements in the current article, and needs to be fixed. Joannamasel (talk) 20:41, 11 November 2013 (UTC)
- Thanks for additions and suggestions! Your expertise on the topic is appreciated. I will make sure we remove any references to neutral mutations being the most common type of mutation. Grabriggs (talk) 01:48, 12 November 2013 (UTC)
- Thank you for the feedback. I've tried to clean up the intro a little so hopefully things are correct and not contradictory.Seanmcaruthers (talk) 04:04, 12 November 2013 (UTC)
Comments from Seth Luty
[edit]Great job, this article is very well written and reads very easy.
- Overall structure and organization is good
- Lots of citations and they are used correctly in the text and look like they are cited correctly in the references
- I love the picture of Darwin, very Epic. But as someone commented on our article you may also want to include a picture of Kimura who also has done a great deal of work in the field
- good use of links, I would link to neutral theory of molecular evolution earlier in the article, for instance in that first section in the third little mini-paragraph - I was expecting it there
- when talking about Synonymous mutation of bases you could expand by talking about wobble theory
- under Identification and measurement of neutrality, Kimura's calculations need to be explained more clearly for the average reader
- the last sentence under Identification and measurement of neutrality about the McDonald-Kreitman test is a little hard to read, the word variation is used to frequently
- possibly expand more on Molecular clocks, or provide an example?
Really good job so far, very strong article. Let me know if you have any questions on my comments. LutyeusMaximus (talk) 16:52, 18 November 2013 (UTC)
- Thanks Seth! I will add in information on wobble theory this week, great idea. Grabriggs (talk) 22:29, 19 November 2013 (UTC)
- I've added a new link to the neutral theory at its first mention. I've also been looking for a Kimura picture but haven't found one that I know is ok with copyright concerns. There's actually one on his article page, but not sure if I can use it legally. I'm going to add wobble theory this evening. Thanks again Seth. Grabriggs (talk) 00:30, 26 November 2013 (UTC)
- Thanks for the great review. I have expanded on Kimura's calculations that I think makes everything a little more understandable. I trimmed up the McDonald-Kreitman section a little and provided a link in external sources to an online application where you can plug sequence data in and run the test. I expanded the molecular clocks section that should show their significance and how they relate to neutral mutation better.Seanmcaruthers (talk) 07:15, 26 November 2013 (UTC)
- Hey I am happy my comments were of help! I hear ya on the finding a legal picture, I had trouble with the same thing with one of our images and I eventually had to just scrap the idea of adding that picture. Also I really like the addition of the amino acid codon table! LutyeusMaximus (talk) 01:59, 4 December 2013 (UTC)
- Thanks for the great review. I have expanded on Kimura's calculations that I think makes everything a little more understandable. I trimmed up the McDonald-Kreitman section a little and provided a link in external sources to an online application where you can plug sequence data in and run the test. I expanded the molecular clocks section that should show their significance and how they relate to neutral mutation better.Seanmcaruthers (talk) 07:15, 26 November 2013 (UTC)
- I've added a new link to the neutral theory at its first mention. I've also been looking for a Kimura picture but haven't found one that I know is ok with copyright concerns. There's actually one on his article page, but not sure if I can use it legally. I'm going to add wobble theory this evening. Thanks again Seth. Grabriggs (talk) 00:30, 26 November 2013 (UTC)
Comments from sefacci (Sarah)
[edit]I'm going to add piecewise to these comments over the next few days since I have time in small increments.
Intro:
- Agree with previous comment that said use the more common neither/nor instead of not/nor combination. My brain stumbled over this a couple of times before comprehending.
- The term fixation is not explained. This would be ok within our class but perhaps not for a larger audience.
- Someone commented on my article talk page that usually the intro doesn't have citations. Perhaps you're getting too in-depth
in the intro, and some of the more detailed points should be moved down.
- It should probably be briefly mentioned how/by what deleterious mutations are removed from the genome.
Sarah Facci (talk) 02:20, 19 November 2013 (UTC)
Types:
- I'd recommend sticking this section before the history and impact ones. It's nice to know what it is before delving into its effects on others things and how it came to be.
- Also, maybe do a "for example, UCU -> UCC is still serine."
- Nice use of chart image, I think that really adds clarity to the section. I think the last 2 sentences about different selective pressures on codon variants is really important too. Good explanation of amino acid substitution. 209.252.247.242 (talk) 00:48, 20 November 2013 (UTC)
History and Impact:
- The history section is very well written. The only thing I'm not sure about is the citation in the first two sentences. It seems like it should be mentioned in the text that this comes from the Origin of Species, but maybe that's just me.
- It's confusing that "implausibly" links to "Haldane's dilemma." Consider mentioning it by name in the text?
- Also be sure you are linking all necessary terms, such as Motoo Kimura, polymorphic, genotype, exons and introns. Remember this is for a more general audience than biologists.
- Be sure you are linking the first mention of "neutral theory of molecular evolution" - right now it's the second.
- I'd like to see any influential scientists mentioned under the "impact" section as well. Overall really good job so far though.
74.47.149.111 (talk) 06:50, 21 November 2013 (UTC)
- Thanks for the suggestions Sarah. I added an example of base substitution, great suggestion! I've also added links to a number of terms and moved the link for neutral theory to the first mention. 174.17.0.230 (talk) 06:36, 25 November 2013 (UTC)
- forgot to log in before my last comment Grabriggs (talk) 06:37, 25 November 2013 (UTC)
- Thanks Sarah. I have reworked the intro, and I believe addressed the concerns and eliminated the citations.Seanmcaruthers (talk) 07:23, 26 November 2013 (UTC)
- Just popped back in to take a look at your progress and I'm impressed! I can see a lot of hard work has gone into this. I don't really have anything else to add at this point. Nice work! There are a couple of style/grammar things that I had to read twice but they are minor and I'm sure will be fixed. Sarah Facci (talk) 05:53, 5 December 2013 (UTC)
- Thanks Sarah. I have reworked the intro, and I believe addressed the concerns and eliminated the citations.Seanmcaruthers (talk) 07:23, 26 November 2013 (UTC)
- forgot to log in before my last comment Grabriggs (talk) 06:37, 25 November 2013 (UTC)
Comments from Keilana
[edit]Hi guys, great job so far. Here are some comments I have for you - please feel free to let me know if you have questions!
- The lead is written at too high a level, you should gloss more things and make it a little bit more accessible, then go into detail in the body of the article.
- You don't need to cite things in the lead that are discussed and cited later in the article because the lead is a summary of the article. Make sure that everything you say in the lead is talked about and cited elsewhere, then you can go ahead and remove those citations.
- Explain what fixation of a mutation and fitness are in the lead.
- Don't use the "and/or" construction, find another way to explain that concept.
- Explain what the neutral theory of molecular evolution is - in general, the lead needs to be expanded and use less technical language.
- Kimura, Duret, Venetianer need PMIDs or dois.
- You did a great job linking important concepts in the lead, but you need more links in the body of the article. For example, Motoo Kimura, polymorphism (biology), intron, and exon should all be linked to.
- It would be great to have a secondary source discussing Darwin's work and Kimura's work. However, you should keep the primary citations.
- The impact section and neutral amino acid substitution section particularly need more links.
- At some point in the article you need to explain the degeneracy of the genetic code.
- "According to the neutral theory (often the nearly neutral)of evolution, the rate of fixation in a population of a neutral mutation will be directly related to the rate of formation of the neutral allele." - this needs clarification (why the parentheses?) and a citation.
- The second paragraph of "Identification and measurement" needs a lot more glossing to be comprehensible to the average reader.
- You definitely need to link to molecular clocks and explain that concept more.
Good luck with the end of your semester! Keilana|Parlez ici 17:52, 20 November 2013 (UTC)
- Thanks Keilana! I have added dois to the articles. I also added a number of links in the article including the ones you suggested. I will be adding something on the degeneracy of the code soon. Grabriggs (talk) 00:15, 24 November 2013 (UTC)
- Thanks for the great review and suggestions. I have reworked the intro and eliminated the citations. I cleared up the "According to the neutral theory...." sentence and added a citation. I explained the variables in Kimura's equation and a little more on what the equation actually means and showed its flaws. The molecular clocks section has been expanded. I hope I am showing just the relevance to neutral mutation, as I think the page on Molecular clocks should be where the heavy details are covered.Seanmcaruthers (talk) 07:30, 26 November 2013 (UTC)
Review by Sridenour
[edit]- The third sentence that starts with ‘If inheritable…’ needs worked on. It seems that perhaps this can be broken down into 2 sentences with the second sentence being altered to be a coherent sentence.
- In the history section, is it possible to wikilink ‘natural theory of molecular evolution’ in the last sentence of the second paragraph.
- The third sentence under Impact on evolutionary theory needs a comma after sequence.
- Great work on the ‘Synonymous mutation of bases’ section. The wording was very informative but do you think you could find a clearer photo. The one here seems a bit blurry but I understand the difficulties of finding suitable images.
- Perhaps you could add an image of a electrophoresis gel to the Identification and measurement of neutrality to break up the long paragraph. But the paragraph overall has great info.
- Reference 16 & 21-why are the author’s names in caps for these references?
In general though really great job…I think the page flows well and has substantial information. Good Job guys. Sridenour (talk) 20:24, 2 December 2013 (UTC)
- Thanks for the input! I think we are only supposed to wikilink the first time a term is used in an article, so we've got the "neutral theory of molecular evolution" linked earlier. I can certainly change this though if I can find something saying that's ok. I updated the picture. You were right, the one we had did not show well and the first one we had was confusing. I think the latest picture is the best and is easy to refer to. We will keep making improvements, thanks again! Grabriggs (talk) 01:59, 3 December 2013 (UTC)
- Hey yes you are correct about redundant wikilinking, sorry I missed the first linking for that. And the new picture looks great. Sridenour (talk) 12:43, 6 December 2013 (UTC)
- Thanks for the very helpful review. I reworked the sentence in the lead and hopefully it is a little more comprehensible. The capitalization of the authors’ names in the references is a product of the import tool from the PMID and DOI numbers. Honestly, I don’t know if I should leave it or change it for consistency.Seanmcaruthers (talk) 07:52, 12 December 2013 (UTC)
- I think it would look better if changed but not 100% on the need for the change so perhaps you can address one of the wiki ambassadors about if this is necessary. Sridenour (talk) 16:15, 12 December 2013 (UTC)
- Thanks for the very helpful review. I reworked the sentence in the lead and hopefully it is a little more comprehensible. The capitalization of the authors’ names in the references is a product of the import tool from the PMID and DOI numbers. Honestly, I don’t know if I should leave it or change it for consistency.Seanmcaruthers (talk) 07:52, 12 December 2013 (UTC)
- Hey yes you are correct about redundant wikilinking, sorry I missed the first linking for that. And the new picture looks great. Sridenour (talk) 12:43, 6 December 2013 (UTC)
A couple more comments from Klortho
[edit]Wow! What a huge difference since the last time I looked at this article. You have done a really fantastic job! It is well organized and very readable. Here are a few more comments that I hope might improve it just a bit more.
- In "Synonymous mutations of bases", I think maybe you have a little too much detail about the "wobble concept". All you should need to do is describe the degeneracy of the code, and link to the relevant article (which you've done) if they want more information. On the other hand, I think it would be good, in that section, to summarize the most common base substitutions that are neutral. For example, summarize that neutral mutations often involve the third base of a codon, and anything else that might be relevant
- Can you find a genetic code chart that uses DNA bases instead of RNA ("T" instead of "U"). It's more familiar to non-biologists, and more appropriate in this article, I think.
- In "Neutral amino acid substitution", more than half of this paragraph is a recapitulation of stuff that you have already talked about, and could be removed. Also, again, I think you need a succinct summary of which types of amino acid substitutes are most often neutral. You have a nice summary of the dependency on location in a protein -- good.
Klortho (talk) 14:52, 8 December 2013 (UTC)
- Thanks again Klortho! I will try to figure out a way to explain degeneracy a little more succinctly. As far as the chart, this is the third one I've tried and each time there seems to be concerns from the editors. I can review each of the concerns and see if I can find a different chart that works. Finally, I can take a look at the AA substitution section and pare it down to take out redundant material. Your input is much appreciated, thanks! Grabriggs (talk) 05:03, 11 December 2013 (UTC)
Comments from Birdy0124
[edit]This is a well-researched article and has lots of good information. Here are some suggestions/comments:
1. Lead section
a. The second sentence “mutations in which natural selection …” may need rewording. Since it is mostly a recap of the first sentence, you probably could cut the sentence out.
b. In the third sentence “If inheritable …”, “replacement of all other alleles of the gene” seems to be out of place (see also comment by Sridenour).
c. Do neutral mutations refer to only germline mutations in the context of this article? Somatic mutations cannot be passed on to progenies. Most of the cancer mutations are somatic. It may be useful to make that clear (although it does not have to in the lead section).
d. The second and third paragraphs are on the same subject. It may flow better to merge them. The first sentence of the third paragraph can be removed. After the last sentence of the second paragraph, you can probably just say “This has led to the development of the neutral …”
2. History
a) Looking through the whole article, you talk about history every time a new concept is introduced. So I am not sure you need a separate section on history. But if you decide to keep the history section, maybe move it toward the end. Most wiki readers want to learn about the subject first before delving into the history.
b) The quote from Charles Darwin shows a “neutral” view on “neutral mutations”. I do not see how one can go from that quote to his view of “change being driven by traits”.
c) Did Charles Darwin first introduce the concept of neutral mutations? If so, it is good to state so.
3. Impact on evolutionary theory
a) Because the article is about “neutral mutation” not “neutral theory of molecular evolution” (wiki already has an article about the latter), it may be better to move this section after you have explained neutral mutation. In fact, it may be better to combine this section with Identification and Measurement Section since all both talk about how the concept of neutral mutations is used to study evolution.
b) “Neutral mutation has become a part of the neutral theory …” Instead of “a part”, should it be “the cornerstone”?
c) The last sentence of the first paragraph paraphrases what was said before and can be deleted.
d) The things you talked about in the second paragraph explain the types and locations of neutral mutations. They are not the “impacts due to neutral mutation”.
4. Type of neutral mutation
a) This should probably be the first section.
b) The “Synonymous mutations” subsection can be condensed to a few sentences (as suggested by another reviewer). Not sure you need the first sentence. Since mutations are at DNA level, it might be good to use a DNA codon table (http://en.wikipedia.org/wiki/DNA_codon_table). The codon table is self-explanatory. Your explanation of the concept of wobble is nice, but it is probably not needed here. However, the point you made about the translational efficiency is important and often overlooked.
c) The neutral amino acid substitution subsection. Based on the title, this subsection should focus on cases where mutations lead to amino acid substitution but the are nevertheless “neutral”. This happens, for example, when substitutions result in amino acids with similar physicochemical property or when mutations involve residues that are not structurally or functionally important (e.g. residues on the surface of the protein).
5. Identification and measurement of neutrality
a) This section has mixed material. Some of the information (e.g. mutations that cause no appreciable effect on gene function etc) should be included in the section “Types of neutral mutations."
b) The focus of this section seems to be the measurement of neutrality in population. It might work if you combine this section with Impact on Evolution Theory section. You probably don’t need to go into too much details. Instead a high level overview of the key concepts will suffice.
6. Molecular clocks
a) “Molecular clocks can be used to measure …” “measure” should be “estimate.”
b) This sentence “The amount of neutral sites in a protein or sequence versus the amount of constrained sites effects the neutral mutation rate in any particular protein or DNA sequenced studied” is very technical. Can you make it simpler for the general audience?
7. References
Great job in finding all these references.
Birdy0124 (talk) 15:36, 9 December 2013 (UTC)
- Thanks Birdy! I've added the chart you linked to in your comments. I've tried three others this semester and each time editors have had concerns, so this is great! You've made a lot of good comments and I will try to address as many as possible this week. Grabriggs (talk) 05:24, 11 December 2013 (UTC)
- Thanks so much for the helpful review. I have reworked the lead taking many of your suggestions. I left some of the material that you recommended leaving out as I think it helps guide the reader to a better idea of what the article covers. I took your suggestions for changes in the molecular clocks section and hopefully things are clearer.Seanmcaruthers (talk) 07:53, 12 December 2013 (UTC)
- I've made some additional edits trying to clarify the Darwin statements, neutral impacts to observations and tried to condense some ideas. Thanks again Grabriggs (talk) 00:36, 13 December 2013 (UTC)
- Thanks so much for the helpful review. I have reworked the lead taking many of your suggestions. I left some of the material that you recommended leaving out as I think it helps guide the reader to a better idea of what the article covers. I took your suggestions for changes in the molecular clocks section and hopefully things are clearer.Seanmcaruthers (talk) 07:53, 12 December 2013 (UTC)
Darwin fetishism
[edit]It is utterly absurd that a page about neutral mutation has an image of Charles Darwin, but not any image of, for instance, Motoo Kimura. Darwin certainly did not originate the idea of neutral mutation. To the extent that he understood mutation and discrete genetics, he rejected their relevance to evolution. The only legitimate context for putting the words "Darwin" and "mutation" in the same sentence is to explain why these two words should not be in the same sentence. Darwin's quoted speculation about variations of no importance is mundane or trivial. There is no evidence that this is the actual source of the ideas of neutrality that began to emerge among biochemists looking at protein molecules in the 1960s. This is Darwin-worship, pure and simple. We do not need more gratuitous Darwin-worship.
This history section should be deleted, or it should be refocused on the molecular revolution, perhaps starting with the work of historian Michael Dietrich. — Preceding unsigned comment added by Dabs (talk • contribs) 16:08, 29 August 2019 (UTC)
- C-Class Molecular Biology articles
- Unknown-importance Molecular Biology articles
- C-Class Genetics articles
- Mid-importance Genetics articles
- WikiProject Genetics articles
- C-Class MCB articles
- High-importance MCB articles
- WikiProject Molecular and Cellular Biology articles
- All WikiProject Molecular Biology pages


