Alternative splicing

Alternative splicing, or alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included within or excluded from the final RNA product of the gene.[1] This means the exons are joined in different combinations, leading to different splice variants. In the case of protein-coding genes, the proteins translated from these splice variants may contain differences in their amino acid sequence and in their biological functions (see Figure).
Biologically relevant alternative splicing occurs as a normal phenomenon in eukaryotes, where it increases the number of proteins that can be encoded by the genome.[1] In humans, it is widely believed that ~95% of multi-exonic genes are alternatively spliced to produce functional alternative products from the same gene[2] but many scientists believe that most of the observed splice variants are due to splicing errors and the actual number of biologically relevant alternatively spliced genes is much lower.[3][4]
Discovery
[edit]Alternative splicing was first observed in 1977.[5][6] The adenovirus produces five primary transcripts early in its infectious cycle, prior to viral DNA replication, and an additional one later, after DNA replication begins. The early primary transcripts continue to be produced after DNA replication begins. The additional primary transcript produced late in infection is large and comes from 5/6 of the 32kb adenovirus genome. This is much larger than any of the individual adenovirus mRNAs present in infected cells. Researchers found that the primary RNA transcript produced by adenovirus type 2 in the late phase was spliced in many different ways, resulting in mRNAs encoding different viral proteins. In addition, the primary transcript contained multiple polyadenylation sites, giving different 3' ends for the processed mRNAs.[7][8][9]
In 1981, the first example of alternative splicing in a transcript from a normal, endogenous gene was characterized.[7] The gene encoding the thyroid hormone calcitonin was found to be alternatively spliced in mammalian cells. The primary transcript from this gene contains 6 exons; the calcitonin mRNA contains exons 1–4, and terminates after a polyadenylation site in exon 4. Another mRNA is produced from this pre-mRNA by skipping exon 4, and includes exons 1–3, 5, and 6. It encodes a protein known as CGRP (calcitonin gene related peptide).[10][11] Examples of alternative splicing in immunoglobin gene transcripts in mammals were also observed in the early 1980s.[7][12]
Since then, many other examples of biologically relevant alternative splicing have been found in eukaryotes.[1] The "record-holder" for alternative splicing is a D. melanogaster gene called Dscam, which could potentially have 38,016 splice variants.[13]
In 2021, it was discovered that the genome of adenovirus type 2, the adenovirus in which alternative splicing was first identified, was able to produce a much greater variety of splice variants than previously thought.[14] By using next generation sequencing technology, researchers were able to update the human adenovirus type 2 transcriptome and document the presence of 904 splice variants produced by the virus through a complex pattern of alternative splicing. Very few of these splice variants have been shown to be functional, a point that the authors raise in their paper.
- "An outstanding question is what roles the menagerie of novel RNAs play or whether they are spurious molecules generated by an overloaded splicing machinery."[14]
Modes
[edit]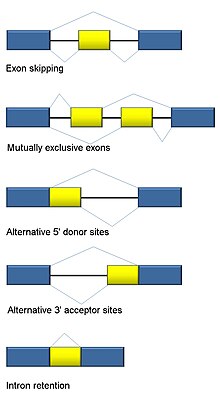
Five basic modes of alternative splicing are generally recognized.[1][2][16][15]
- Exon skipping or cassette exon: in this case, an exon may be spliced out of the primary transcript or retained. This is the most common mode in mammalian pre-mRNAs.[15]
- Mutually exclusive exons: One of two exons is retained in mRNAs after splicing, but not both.
- Alternative donor site: An alternative 5' splice junction (donor site) is used, changing the 3' boundary of the upstream exon.
- Alternative acceptor site: An alternative 3' splice junction (acceptor site) is used, changing the 5' boundary of the downstream exon.
- Intron retention: A sequence may be spliced out as an intron or simply retained. This is distinguished from exon skipping because the retained sequence is not flanked by introns. If the retained intron is in the coding region, the intron must encode amino acids in frame with the neighboring exons, or a stop codon or a shift in the reading frame will cause the protein to be non-functional. This is the rarest mode in mammals but the most common in plants.[15][17]
In addition to these primary modes of alternative splicing, there are two other main mechanisms by which different mRNAs may be generated from the same gene; multiple promoters and multiple polyadenylation sites. Use of multiple promoters is properly described as a transcriptional regulation mechanism rather than alternative splicing; by starting transcription at different points, transcripts with different 5'-most exons can be generated. At the other end, multiple polyadenylation sites provide different 3' end points for the transcript. Both of these mechanisms are found in combination with alternative splicing and provide additional variety in mRNAs derived from a gene.[1][16]
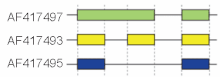
These modes describe basic splicing mechanisms, but may be inadequate to describe complex splicing events. For instance, the figure to the right shows 3 spliceforms from the mouse hyaluronidase 3 gene. Comparing the exonic structure shown in the first line (green) with the one in the second line (yellow) shows intron retention, whereas the comparison between the second and the third spliceform (yellow vs. blue) exhibits exon skipping. A model nomenclature to uniquely designate all possible splicing patterns has recently been proposed.[15]
Mechanisms
[edit]General splicing mechanism
[edit]
When the pre-mRNA has been transcribed from the DNA, it includes several introns and exons. (In nematodes, the mean is 4–5 exons and introns; in the fruit fly Drosophila there can be more than 100 introns and exons in one transcribed pre-mRNA.) The exons to be retained in the mRNA are determined during the splicing process. The regulation and selection of splice sites are done by trans-acting splicing activator and splicing repressor proteins as well as cis-acting elements within the pre-mRNA itself such as exonic splicing enhancers and exonic splicing silencers.
The typical eukaryotic nuclear intron has consensus sequences defining important regions. Each intron has the sequence GU at its 5' end. Near the 3' end there is a branch site. The nucleotide at the branchpoint is always an A; the consensus around this sequence varies somewhat. In humans the branch site consensus sequence is yUnAy.[18] The branch site is followed by a series of pyrimidines – the polypyrimidine tract – then by AG at the 3' end.[16]
Splicing of mRNA is performed by an RNA and protein complex known as the spliceosome, containing snRNPs designated U1, U2, U4, U5, and U6 (U3 is not involved in mRNA splicing).[19] U1 binds to the 5' GU and U2, with the assistance of the U2AF protein factors, binds to the branchpoint A within the branch site. The complex at this stage is known as the spliceosome A complex. Formation of the A complex is usually the key step in determining the ends of the intron to be spliced out, and defining the ends of the exon to be retained.[16] (The U nomenclature derives from their high uridine content).
The U4,U5,U6 complex binds, and U6 replaces the U1 position. U1 and U4 leave. The remaining complex then performs two transesterification reactions. In the first transesterification, 5' end of the intron is cleaved from the upstream exon and joined to the branch site A by a 2',5'-phosphodiester linkage. In the second transesterification, the 3' end of the intron is cleaved from the downstream exon, and the two exons are joined by a phosphodiester bond. The intron is then released in lariat form and degraded.[1]
Regulatory elements and proteins
[edit]
Splicing is regulated by trans-acting proteins (repressors and activators) and corresponding cis-acting regulatory sites (silencers and enhancers) on the pre-mRNA. However, as part of the complexity of alternative splicing, it is noted that the effects of a splicing factor are frequently position-dependent. That is, a splicing factor that serves as a splicing activator when bound to an intronic enhancer element may serve as a repressor when bound to its splicing element in the context of an exon, and vice versa.[20] The secondary structure of the pre-mRNA transcript also plays a role in regulating splicing, such as by bringing together splicing elements or by masking a sequence that would otherwise serve as a binding element for a splicing factor.[21][22] Together, these elements form a "splicing code" that governs how splicing will occur under different cellular conditions.[23][24]
There are two major types of cis-acting RNA sequence elements present in pre-mRNAs and they have corresponding trans-acting RNA-binding proteins. Splicing silencers are sites to which splicing repressor proteins bind, reducing the probability that a nearby site will be used as a splice junction. These can be located in the intron itself (intronic splicing silencers, ISS) or in a neighboring exon (exonic splicing silencers, ESS). They vary in sequence, as well as in the types of proteins that bind to them. The majority of splicing repressors are heterogeneous nuclear ribonucleoproteins (hnRNPs) such as hnRNPA1 and polypyrimidine tract binding protein (PTB).[16][23] Splicing enhancers are sites to which splicing activator proteins bind, increasing the probability that a nearby site will be used as a splice junction. These also may occur in the intron (intronic splicing enhancers, ISE) or exon (exonic splicing enhancers, ESE). Most of the activator proteins that bind to ISEs and ESEs are members of the SR protein family. Such proteins contain RNA recognition motifs and arginine and serine-rich (RS) domains.[16][23]
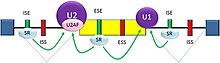
In general, the determinants of splicing work in an inter-dependent manner that depends on context, so that the rules governing how splicing is regulated form a splicing code.[24] The presence of a particular cis-acting RNA sequence element may increase the probability that a nearby site will be spliced in some cases, but decrease the probability in other cases, depending on context. The context within which regulatory elements act includes cis-acting context that is established by the presence of other RNA sequence features, and trans-acting context that is established by cellular conditions. For example, some cis-acting RNA sequence elements influence splicing only if multiple elements are present in the same region so as to establish context. As another example, a cis-acting element can have opposite effects on splicing, depending on which proteins are expressed in the cell (e.g., neuronal versus non-neuronal PTB). The adaptive significance of splicing silencers and enhancers is attested by studies showing that there is strong selection in human genes against mutations that produce new silencers or disrupt existing enhancers.[25][26]
Examples
[edit]Exon skipping: Drosophila dsx
[edit]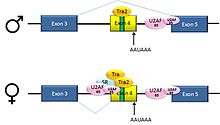
Pre-mRNAs from the D. melanogaster gene dsx contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a polyadenylation signal in exon 4 causes cleavage of the mRNA at that point. The resulting mRNA is a transcriptional regulatory protein required for female development.[27]
This is an example of exon skipping. The intron upstream from exon 4 has a polypyrimidine tract that doesn't match the consensus sequence well, so that U2AF proteins bind poorly to it without assistance from splicing activators. This 3' splice acceptor site is therefore not used in males. Females, however, produce the splicing activator Transformer (Tra) (see below). The SR protein Tra2 is produced in both sexes and binds to an ESE in exon 4; if Tra is present, it binds to Tra2 and, along with another SR protein, forms a complex that assists U2AF proteins in binding to the weak polypyrimidine tract. U2 is recruited to the associated branchpoint, and this leads to inclusion of exon 4 in the mRNA.[27][28]
Alternative acceptor sites: Drosophila Transformer
[edit]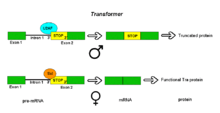
Pre-mRNAs of the Transformer (Tra) gene of Drosophila melanogaster undergo alternative splicing via the alternative acceptor site mode. The gene Tra encodes a protein that is expressed only in females. The primary transcript of this gene contains an intron with two possible acceptor sites. In males, the upstream acceptor site is used. This causes a longer version of exon 2 to be included in the processed transcript, including an early stop codon. The resulting mRNA encodes a truncated protein product that is inactive. Females produce the master sex determination protein Sex lethal (Sxl). The Sxl protein is a splicing repressor that binds to an ISS in the RNA of the Tra transcript near the upstream acceptor site, preventing U2AF protein from binding to the polypyrimidine tract. This prevents the use of this junction, shifting the spliceosome binding to the downstream acceptor site. Splicing at this point bypasses the stop codon, which is excised as part of the intron. The resulting mRNA encodes an active Tra protein, which itself is a regulator of alternative splicing of other sex-related genes (see dsx above).[1]
Exon definition: Fas receptor
[edit]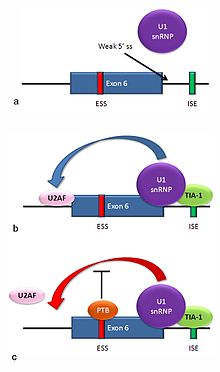
Multiple isoforms of the Fas receptor protein are produced by alternative splicing. Two normally occurring isoforms in humans are produced by an exon-skipping mechanism. An mRNA including exon 6 encodes the membrane-bound form of the Fas receptor, which promotes apoptosis, or programmed cell death. Increased expression of Fas receptor in skin cells chronically exposed to the sun, and absence of expression in skin cancer cells, suggests that this mechanism may be important in elimination of pre-cancerous cells in humans.[29] If exon 6 is skipped, the resulting mRNA encodes a soluble Fas protein that does not promote apoptosis. The inclusion or skipping of the exon depends on two antagonistic proteins, TIA-1 and polypyrimidine tract-binding protein (PTB).
- The 5' donor site in the intron downstream from exon 6 in the pre-mRNA has a weak agreement with the consensus sequence, and is not bound usually by the U1 snRNP. If U1 does not bind, the exon is skipped (see "a" in accompanying figure).
- Binding of TIA-1 protein to an intronic splicing enhancer site stabilizes binding of the U1 snRNP.[16] The resulting 5' donor site complex assists in binding of the splicing factor U2AF to the 3' splice site upstream of the exon, through a mechanism that is not yet known (see b).[30]
- Exon 6 contains a pyrimidine-rich exonic splicing silencer, ure6, where PTB can bind. If PTB binds, it inhibits the effect of the 5' donor complex on the binding of U2AF to the acceptor site, resulting in exon skipping (see c).
This mechanism is an example of exon definition in splicing. A spliceosome assembles on an intron, and the snRNP subunits fold the RNA so that the 5' and 3' ends of the intron are joined. However, recently studied examples such as this one show that there are also interactions between the ends of the exon. In this particular case, these exon definition interactions are necessary to allow the binding of core splicing factors prior to assembly of the spliceosomes on the two flanking introns.[30]
Repressor-activator competition: HIV-1 tat exon 2
[edit]
HIV, the retrovirus that causes AIDS in humans, produces a single primary RNA transcript, which is alternatively spliced in multiple ways to produce over 40 different mRNAs.[31] Equilibrium among differentially spliced transcripts provides multiple mRNAs encoding different products that are required for viral multiplication.[32] One of the differentially spliced transcripts contains the tat gene, in which exon 2 is a cassette exon that may be skipped or included. The inclusion of tat exon 2 in the RNA is regulated by competition between the splicing repressor hnRNP A1 and the SR protein SC35. Within exon 2 an exonic splicing silencer sequence (ESS) and an exonic splicing enhancer sequence (ESE) overlap. If A1 repressor protein binds to the ESS, it initiates cooperative binding of multiple A1 molecules, extending into the 5' donor site upstream of exon 2 and preventing the binding of the core splicing factor U2AF35 to the polypyrimidine tract. If SC35 binds to the ESE, it prevents A1 binding and maintains the 5' donor site in an accessible state for assembly of the spliceosome. Competition between the activator and repressor ensures that both mRNA types (with and without exon 2) are produced.[31]
Adaptive significance
[edit]Genuine alternative splicing occurs in both protein-coding genes and non-coding genes to produce multiple products (proteins or non-coding RNAs). External information is needed in order to decide which product is made, given a DNA sequence and the initial transcript. Since the methods of regulation are inherited, this provides novel ways for mutations to affect gene expression.[33]
Alternative splicing may provide evolutionary flexibility. A single point mutation may cause a given exon to be occasionally excluded or included from a transcript during splicing, allowing production of a new protein isoform without loss of the original protein.[1] Studies have identified intrinsically disordered regions (see Intrinsically unstructured proteins) as enriched in the non-constitutive exons[34] suggesting that protein isoforms may display functional diversity due to the alteration of functional modules within these regions. Such functional diversity achieved by isoforms is reflected by their expression patterns and can be predicted by machine learning approaches.[35][36] Comparative studies indicate that alternative splicing preceded multicellularity in evolution, and suggest that this mechanism might have been co-opted to assist in the development of multicellular organisms.[37]
Research based on the Human Genome Project and other genome sequencing has shown that humans have only about 30% more genes than the roundworm Caenorhabditis elegans, and only about twice as many as the fly Drosophila melanogaster. This finding led to speculation that the perceived greater complexity of humans, or vertebrates generally, might be due to higher rates of alternative splicing in humans than are found in invertebrates.[38][39] However, a study on samples of 100,000 expressed sequence tags (EST) each from human, mouse, rat, cow, fly (D. melanogaster), worm (C. elegans), and the plant Arabidopsis thaliana found no large differences in frequency of alternatively spliced genes among humans and any of the other animals tested.[40] Another study, however, proposed that these results were an artifact of the different numbers of ESTs available for the various organisms. When they compared alternative splicing frequencies in random subsets of genes from each organism, the authors concluded that vertebrates do have higher rates of alternative splicing than invertebrates.[41]
Disease
[edit]Changes in the RNA processing machinery may lead to mis-splicing of multiple transcripts, while single-nucleotide alterations in splice sites or cis-acting splicing regulatory sites may lead to differences in splicing of a single gene, and thus in the mRNA produced from a mutant gene's transcripts. A study in 2005 involving probabilistic analyses indicated that greater than 60% of human disease-causing mutations affect splicing rather than directly affecting coding sequences.[42] A more recent study indicates that one-third of all hereditary diseases are likely to have a splicing component.[20] Regardless of exact percentage, a number of splicing-related diseases do exist.[43] As described below, a prominent example of splicing-related diseases is cancer.
Abnormally spliced mRNAs are also found in a high proportion of cancerous cells.[44][45][46] Combined RNA-Seq and proteomics analyses have revealed striking differential expression of splice isoforms of key proteins in important cancer pathways.[47] It is not always clear whether such aberrant patterns of splicing contribute to the cancerous growth, or are merely consequence of cellular abnormalities associated with cancer. For certain types of cancer, like in colorectal and prostate, the number of splicing errors per cancer has been shown to vary greatly between individual cancers, a phenomenon referred to as transcriptome instability.[48][49] Transcriptome instability has further been shown to correlate grealty with reduced expression level of splicing factor genes. Mutation of DNMT3A has been demonstrated to contribute to hematologic malignancies, and that DNMT3A-mutated cell lines exhibit transcriptome instability as compared to their isogenic wildtype counterparts.[50]
In fact, there is actually a reduction of alternative splicing in cancerous cells compared to normal ones, and the types of splicing differ; for instance, cancerous cells show higher levels of intron retention than normal cells, but lower levels of exon skipping.[51] Some of the differences in splicing in cancerous cells may be due to the high frequency of somatic mutations in splicing factor genes,[46] and some may result from changes in phosphorylation of trans-acting splicing factors.[33] Others may be produced by changes in the relative amounts of splicing factors produced; for instance, breast cancer cells have been shown to have increased levels of the splicing factor SF2/ASF.[52] One study found that a relatively small percentage (383 out of over 26000) of alternative splicing variants were significantly higher in frequency in tumor cells than normal cells, suggesting that there is a limited set of genes which, when mis-spliced, contribute to tumor development.[53] It is believed however that the deleterious effects of mis-spliced transcripts are usually safeguarded and eliminated by a cellular posttranscriptional quality control mechanism termed nonsense-mediated mRNA decay [NMD].[54]
One example of a specific splicing variant associated with cancers is in one of the human DNMT genes. Three DNMT genes encode enzymes that add methyl groups to DNA, a modification that often has regulatory effects. Several abnormally spliced DNMT3B mRNAs are found in tumors and cancer cell lines. In two separate studies, expression of two of these abnormally spliced mRNAs in mammalian cells caused changes in the DNA methylation patterns in those cells. Cells with one of the abnormal mRNAs also grew twice as fast as control cells, indicating a direct contribution to tumor development by this product.[33]
Another example is the Ron (MST1R) proto-oncogene. An important property of cancerous cells is their ability to move and invade normal tissue. Production of an abnormally spliced transcript of Ron has been found to be associated with increased levels of the SF2/ASF in breast cancer cells. The abnormal isoform of the Ron protein encoded by this mRNA leads to cell motility.[52]
Overexpression of a truncated splice variant of the FOSB gene – ΔFosB – in a specific population of neurons in the nucleus accumbens has been identified as the causal mechanism involved in the induction and maintenance of an addiction to drugs and natural rewards.[55][56][57][58]
Recent provocative studies point to a key function of chromatin structure and histone modifications in alternative splicing regulation. These insights suggest that epigenetic regulation determines not only what parts of the genome are expressed but also how they are spliced.[59]
Genome-scale (transcriptome-wide) analysis
[edit]Transcriptome-wide analysis of alternative splicing is typically performed by high-throughput RNA-sequencing. Most commonly, by short-read sequencing, such as by Illumina instrumentation. But even more informative, by long-read sequencing, such as by Nanopore or PacBio instrumentation. Transcriptome-wide analyses can for example be used to measure the amount of deviating alternative splicing, such as in a cancer cohort.[60]
Deep sequencing technologies have been used to conduct genome-wide analyses of both unprocessed and processed mRNAs; thus providing insights into alternative splicing. For example, results from use of deep sequencing indicate that, in humans, an estimated 95% of transcripts from multiexon genes undergo alternative splicing, with a number of pre-mRNA transcripts spliced in a tissue-specific manner.[2] Functional genomics and computational approaches based on multiple instance learning have also been developed to integrate RNA-seq data to predict functions for alternatively spliced isoforms.[36] Deep sequencing has also aided in the in vivo detection of the transient lariats that are released during splicing, the determination of branch site sequences, and the large-scale mapping of branchpoints in human pre-mRNA transcripts.[61]
More historically, alternatively spliced transcripts have been found by comparing EST sequences, but this requires sequencing of very large numbers of ESTs. Most EST libraries come from a very limited number of tissues, so tissue-specific splice variants are likely to be missed in any case. High-throughput approaches to investigate splicing have, however, been developed, such as: DNA microarray-based analyses, RNA-binding assays, and deep sequencing. These methods can be used to screen for polymorphisms or mutations in or around splicing elements that affect protein binding. When combined with splicing assays, including in vivo reporter gene assays, the functional effects of polymorphisms or mutations on the splicing of pre-mRNA transcripts can then be analyzed.[20][23][62]
In microarray analysis, arrays of DNA fragments representing individual exons (e.g. Affymetrix exon microarray) or exon/exon boundaries (e.g. arrays from ExonHit or Jivan) have been used. The array is then probed with labeled cDNA from tissues of interest. The probe cDNAs bind to DNA from the exons that are included in mRNAs in their tissue of origin, or to DNA from the boundary where two exons have been joined. This can reveal the presence of particular alternatively spliced mRNAs.[63]
CLIP (Cross-linking and immunoprecipitation) uses UV radiation to link proteins to RNA molecules in a tissue during splicing. A trans-acting splicing regulatory protein of interest is then precipitated using specific antibodies. When the RNA attached to that protein is isolated and cloned, it reveals the target sequences for that protein.[64] Another method for identifying RNA-binding proteins and mapping their binding to pre-mRNA transcripts is "Microarray Evaluation of Genomic Aptamers by shift (MEGAshift)".net[65] This method involves an adaptation of the "Systematic Evolution of Ligands by Exponential Enrichment (SELEX)" method[66] together with a microarray-based readout. Use of the MEGAshift method has provided insights into the regulation of alternative splicing by allowing for the identification of sequences in pre-mRNA transcripts surrounding alternatively spliced exons that mediate binding to different splicing factors, such as ASF/SF2 and PTB.[67] This approach has also been used to aid in determining the relationship between RNA secondary structure and the binding of splicing factors.[22]
Use of reporter assays makes it possible to find the splicing proteins involved in a specific alternative splicing event by constructing reporter genes that will express one of two different fluorescent proteins depending on the splicing reaction that occurs. This method has been used to isolate mutants affecting splicing and thus to identify novel splicing regulatory proteins inactivated in those mutants.[64]
Recent advancements in protein structure prediction have facilitated the development of new tools for genome annotation and alternative splicing anlaysis. For instance, isoform.io, a platform guided by protein structure predictions, has evaluated hundreds of thousands of isoforms of human protein-coding genes assembled from numerous RNA sequencing experiments across a variety of human tissues. This comprehensive analysis has led to the identification of numerous isoforms with more confidently predicted structure and potentially superior function compared to canonical isoforms in the latest human gene database. By integrating structural predictions with expression and evolutionary evidence, this approach has demonstrated the potential of protein structure prediction as a tool for refining the annotation of the human genome.[68]
Databases
[edit]There is a collection of alternative splicing databases.[69][70][71] These databases are useful for finding genes having pre-mRNAs undergoing alternative splicing and alternative splicing events or to study the functional impact of alternative splicing.
- AspicDB database
- Intronerator database
- ProSAS database
See also
[edit]References
[edit]- ^ a b c d e f g h Black DL (2003). "Mechanisms of alternative pre-messenger RNA splicing" (PDF). Annual Review of Biochemistry. 72 (1): 291–336. doi:10.1146/annurev.biochem.72.121801.161720. PMID 12626338. S2CID 23576288.
- ^ a b c Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ (December 2008). "Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing". Nature Genetics. 40 (12): 1413–5. doi:10.1038/ng.259. PMID 18978789. S2CID 9228930.
- ^ Bhuiyan SA, Ly S, Phan M, Huntington B, Hogan E, Liu CC, Liu J, Pavlidis P (2018). "Systematic evaluation of isoform function in literature reports of alternative splicing". BMC Genomics. 19 (1): 637. doi:10.1186/s12864-018-5013-2. PMC 6114036. PMID 30153812. S2CID 52113302.
- ^ Tress ML, Abascal F, Valencia A (2017). "Alternative splicing may not be the key to proteome complexity". Trends in Biochemical Sciences. 42 (2): 98–110. doi:10.1016/j.tibs.2016.08.008. PMC 6526280. PMID 27712956.
- ^ Chow LT, Gelinas RE, Broker TR, Roberts RJ (September 1977). "An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA". Cell. 12 (1): 1–8. doi:10.1016/0092-8674(77)90180-5. PMID 902310. S2CID 2099968.
- ^ Berget SM, Moore C, Sharp PA (August 1977). "Spliced segments at the 5' terminus of adenovirus 2 late mRNA". Proceedings of the National Academy of Sciences of the United States of America. 74 (8): 3171–5. Bibcode:1977PNAS...74.3171B. doi:10.1073/pnas.74.8.3171. PMC 431482. PMID 269380.
- ^ a b c Leff SE, Rosenfeld MG, Evans RM (1986). "Complex transcriptional units: diversity in gene expression by alternative RNA processing". Annual Review of Biochemistry. 55 (1): 1091–117. doi:10.1146/annurev.bi.55.070186.005303. PMID 3017190.
- ^ Chow LT, Broker TR (October 1978). "The spliced structures of adenovirus 2 fiber message and the other late mRNAs". Cell. 15 (2): 497–510. doi:10.1016/0092-8674(78)90019-3. PMID 719751. S2CID 44642349.
- ^ Nevins JR, Darnell JE (December 1978). "Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing". Cell. 15 (4): 1477–93. doi:10.1016/0092-8674(78)90071-5. PMID 729004. S2CID 39704416.
- ^ Rosenfeld MG, Amara SG, Roos BA, Ong ES, Evans RM (March 1981). "Altered expression of the calcitonin gene associated with RNA polymorphism". Nature. 290 (5801): 63–5. Bibcode:1981Natur.290...63R. doi:10.1038/290063a0. PMID 7207587. S2CID 4318349.
- ^ Rosenfeld MG, Lin CR, Amara SG, Stolarsky L, Roos BA, Ong ES, Evans RM (March 1982). "Calcitonin mRNA polymorphism: peptide switching associated with alternative RNA splicing events". Proceedings of the National Academy of Sciences of the United States of America. 79 (6): 1717–21. Bibcode:1982PNAS...79.1717R. doi:10.1073/pnas.79.6.1717. PMC 346051. PMID 6952224.
- ^ Maki R, Roeder W, Traunecker A, Sidman C, Wabl M, Raschke W, Tonegawa S (May 1981). "The role of DNA rearrangement and alternative RNA processing in the expression of immunoglobulin delta genes". Cell. 24 (2): 353–65. doi:10.1016/0092-8674(81)90325-1. PMID 6786756. S2CID 13208589.
- ^ Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, et al. (June 2000). "Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity". Cell. 101 (6): 671–84. doi:10.1016/S0092-8674(00)80878-8. PMID 10892653. S2CID 13829976.
- ^ a b Westergren Jakobsson A, Segerman B, Wallerman O, Lind SB, Zhao H, Rubin CJ, et al. (November 2020). "The Human Adenovirus Type 2 Transcriptome: An Amazing Complexity of Alternatively Spliced mRNAs". Journal of Virology. 95 (4). doi:10.1128/JVI.01869-20. PMC 7851563. PMID 33239457.
- ^ a b c d e Sammeth M, Foissac S, Guigó R (August 2008). Brent MR (ed.). "A general definition and nomenclature for alternative splicing events". PLOS Computational Biology. 4 (8): e1000147. Bibcode:2008PLSCB...4E0147S. doi:10.1371/journal.pcbi.1000147. PMC 2467475. PMID 18688268.
- ^ a b c d e f g h Matlin AJ, Clark F, Smith CW (May 2005). "Understanding alternative splicing: towards a cellular code". Nature Reviews. Molecular Cell Biology. 6 (5): 386–98. doi:10.1038/nrm1645. PMID 15956978. S2CID 14883495.
- ^ Ner-Gaon, Hadas; Halachmi, Ronit; Savaldi-Goldstein, Sigal; Rubin, Eitan; Ophir, Ron; Fluhr, Robert (2004). "Intron retention is a major phenomenon in alternative splicing in Arabidopsis". The Plant Journal. 39 (6): 877–885. doi:10.1111/j.1365-313X.2004.02172.x. ISSN 1365-313X. PMID 15341630.
- ^ Gao K, Masuda A, Matsuura T, Ohno K (April 2008). "Human branch point consensus sequence is yUnAy". Nucleic Acids Research. 36 (7): 2257–67. doi:10.1093/nar/gkn073. PMC 2367711. PMID 18285363.
- ^ Clark D (2005). Molecular biology. Amsterdam: Elsevier Academic Press. ISBN 978-0-12-175551-5.
- ^ a b c Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG (July 2011). "Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes". Proceedings of the National Academy of Sciences of the United States of America. 108 (27): 11093–8. Bibcode:2011PNAS..10811093H. doi:10.1073/pnas.1101135108. PMC 3131313. PMID 21685335.
- ^ Warf MB, Berglund JA (March 2010). "Role of RNA structure in regulating pre-mRNA splicing". Trends in Biochemical Sciences. 35 (3): 169–78. doi:10.1016/j.tibs.2009.10.004. PMC 2834840. PMID 19959365.
- ^ a b Reid DC, Chang BL, Gunderson SI, Alpert L, Thompson WA, Fairbrother WG (December 2009). "Next-generation SELEX identifies sequence and structural determinants of splicing factor binding in human pre-mRNA sequence". RNA. 15 (12): 2385–97. doi:10.1261/rna.1821809. PMC 2779669. PMID 19861426.
- ^ a b c d Wang Z, Burge CB (May 2008). "Splicing regulation: from a parts list of regulatory elements to an integrated splicing code" (Free full text). RNA. 14 (5): 802–13. doi:10.1261/rna.876308. PMC 2327353. PMID 18369186.
- ^ a b Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, et al. (May 2010). "Deciphering the splicing code". Nature. 465 (7294): 53–9. Bibcode:2010Natur.465...53B. doi:10.1038/nature09000. PMID 20445623. S2CID 2398858.
- ^ Ke S, Zhang XH, Chasin LA (April 2008). "Positive selection acting on splicing motifs reflects compensatory evolution". Genome Research. 18 (4): 533–43. doi:10.1101/gr.070268.107. PMC 2279241. PMID 18204002.
- ^ Fairbrother WG, Holste D, Burge CB, Sharp PA (September 2004). "Single nucleotide polymorphism-based validation of exonic splicing enhancers". PLOS Biology. 2 (9): E268. doi:10.1371/journal.pbio.0020268. PMC 514884. PMID 15340491.
- ^ a b Lynch KW, Maniatis T (August 1996). "Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer". Genes & Development. 10 (16): 2089–101. doi:10.1101/gad.10.16.2089. PMID 8769651.
- ^ Graveley BR, Hertel KJ, Maniatis T (June 2001). "The role of U2AF35 and U2AF65 in enhancer-dependent splicing". RNA. 7 (6): 806–18. doi:10.1017/S1355838201010317. PMC 1370132. PMID 11421359.
- ^ Filipowicz E, Adegboyega P, Sanchez RL, Gatalica Z (February 2002). "Expression of CD95 (Fas) in sun-exposed human skin and cutaneous carcinomas". Cancer. 94 (3): 814–9. doi:10.1002/cncr.10277. PMID 11857317. S2CID 23772719.
- ^ a b Izquierdo JM, Majós N, Bonnal S, Martínez C, Castelo R, Guigó R, et al. (August 2005). "Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition". Molecular Cell. 19 (4): 475–84. doi:10.1016/j.molcel.2005.06.015. PMID 16109372.
- ^ a b Zahler AM, Damgaard CK, Kjems J, Caputi M (March 2004). "SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing". The Journal of Biological Chemistry. 279 (11): 10077–84. doi:10.1074/jbc.M312743200. PMID 14703516.
- ^ Jacquenet S, Méreau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C (November 2001). "A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H". The Journal of Biological Chemistry. 276 (44): 40464–75. doi:10.1074/jbc.M104070200. PMID 11526107.
- ^ a b c Fackenthal JD, Godley LA (2008). "Aberrant RNA splicing and its functional consequences in cancer cells" (Free full text). Disease Models & Mechanisms. 1 (1): 37–42. doi:10.1242/dmm.000331. PMC 2561970. PMID 19048051.
- ^ Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, et al. (May 2006). "Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms". Proceedings of the National Academy of Sciences of the United States of America. 103 (22): 8390–5. Bibcode:2006PNAS..103.8390R. doi:10.1073/pnas.0507916103. PMC 1482503. PMID 16717195.
- ^ Li HD, Menon R, Omenn GS, Guan Y (August 2014). "The emerging era of genomic data integration for analyzing splice isoform function". Trends in Genetics. 30 (8): 340–7. doi:10.1016/j.tig.2014.05.005. PMC 4112133. PMID 24951248.
- ^ a b Eksi R, Li HD, Menon R, Wen Y, Omenn GS, Kretzler M, Guan Y (Nov 2013). "Systematically differentiating functions for alternatively spliced isoforms through integrating RNA-seq data". PLOS Computational Biology. 9 (11): e1003314. Bibcode:2013PLSCB...9E3314E. doi:10.1371/journal.pcbi.1003314. PMC 3820534. PMID 24244129.
- ^ Irimia M, Rukov JL, Penny D, Roy SW (October 2007). "Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing". BMC Evolutionary Biology. 7 (1): 188. Bibcode:2007BMCEE...7..188I. doi:10.1186/1471-2148-7-188. PMC 2082043. PMID 17916237.
- ^ Ewing B, Green P (June 2000). "Analysis of expressed sequence tags indicates 35,000 human genes". Nature Genetics. 25 (2): 232–4. doi:10.1038/76115. PMID 10835644. S2CID 19165121.
- ^ Roest Crollius H, Jaillon O, Bernot A, Dasilva C, Bouneau L, Fischer C, et al. (June 2000). "Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence". Nature Genetics. 25 (2): 235–8. doi:10.1038/76118. PMID 10835645. S2CID 44052050.
- ^ Brett D, Pospisil H, Valcárcel J, Reich J, Bork P (January 2002). "Alternative splicing and genome complexity". Nature Genetics. 30 (1): 29–30. doi:10.1038/ng803. PMID 11743582. S2CID 2724843.
- ^ Kim E, Magen A, Ast G (2006). "Different levels of alternative splicing among eukaryotes". Nucleic Acids Research. 35 (1): 125–31. doi:10.1093/nar/gkl924. PMC 1802581. PMID 17158149.
- ^ López-Bigas N, Audit B, Ouzounis C, Parra G, Guigó R (March 2005). "Are splicing mutations the most frequent cause of hereditary disease?". FEBS Letters. 579 (9): 1900–3. Bibcode:2005FEBSL.579.1900L. doi:10.1016/j.febslet.2005.02.047. PMID 15792793. S2CID 30174458.
- ^ Ward AJ, Cooper TA (January 2010). "The pathobiology of splicing". The Journal of Pathology. 220 (2): 152–63. doi:10.1002/path.2649. PMC 2855871. PMID 19918805.
- ^ Skotheim RI, Nees M (2007). "Alternative splicing in cancer: noise, functional, or systematic?". The International Journal of Biochemistry & Cell Biology. 39 (7–8): 1432–49. doi:10.1016/j.biocel.2007.02.016. PMID 17416541.
- ^ He C, Zhou F, Zuo Z, Cheng H, Zhou R (2009). Bauer JA (ed.). "A global view of cancer-specific transcript variants by subtractive transcriptome-wide analysis". PLOS ONE. 4 (3): e4732. Bibcode:2009PLoSO...4.4732H. doi:10.1371/journal.pone.0004732. PMC 2648985. PMID 19266097.
- ^ a b Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI (May 2016). "Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes". Oncogene. 35 (19): 2413–27. doi:10.1038/onc.2015.318. PMID 26300000. S2CID 22943729.
- ^ Omenn GS, Guan Y, Menon R (July 2014). "A new class of protein cancer biomarker candidates: differentially expressed splice variants of ERBB2 (HER2/neu) and ERBB1 (EGFR) in breast cancer cell lines". Journal of Proteomics. 107: 103–12. doi:10.1016/j.jprot.2014.04.012. PMC 4123867. PMID 24802673.
- ^ Sveen A, Johannessen B, Teixeira MR, Lothe RA, Skotheim RI (August 2014). "Transcriptome instability as a molecular pan-cancer characteristic of carcinomas". BMC Genomics. 15 (1): 672. doi:10.1186/1471-2164-15-672. PMC 3219073. PMID 25109687.
- ^ Sveen A, Agesen TH, Nesbakken A, Rognum TO, Lothe RA, Skotheim RI (May 2011). "Transcriptome instability in colorectal cancer identified by exon microarray analyses: Associations with splicing factor expression levels and patient survival". Genome Medicine. 3 (5): 32. doi:10.1186/gm248. PMC 4137096. PMID 21619627.
- ^ Banaszak LG, Giudice V, Zhao X, Wu Z, Gao S, Hosokawa K, et al. (March 2018). "Abnormal RNA splicing and genomic instability after induction of DNMT3A mutations by CRISPR/Cas9 gene editing". Blood Cells, Molecules & Diseases. 69: 10–22. doi:10.1016/j.bcmd.2017.12.002. PMC 6728079. PMID 29324392.
- ^ Kim E, Goren A, Ast G (January 2008). "Insights into the connection between cancer and alternative splicing". Trends in Genetics. 24 (1): 7–10. doi:10.1016/j.tig.2007.10.001. PMID 18054115.
- ^ a b Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, et al. (December 2005). "Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene". Molecular Cell. 20 (6): 881–90. doi:10.1016/j.molcel.2005.10.026. PMID 16364913.
- ^ Hui L, Zhang X, Wu X, Lin Z, Wang Q, Li Y, Hu G (April 2004). "Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment". Oncogene. 23 (17): 3013–23. doi:10.1038/sj.onc.1207362. PMID 15048092.
- ^ Danckwardt S, Neu-Yilik G, Thermann R, Frede U, Hentze MW, Kulozik AE (March 2002). "Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay". Blood. 99 (5): 1811–6. doi:10.1182/blood.V99.5.1811. PMID 11861299. S2CID 17128174.
- ^ Nestler EJ (December 2013). "Cellular basis of memory for addiction". Dialogues in Clinical Neuroscience. 15 (4): 431–43. doi:10.31887/DCNS.2013.15.4/enestler. PMC 3898681. PMID 24459410.
DESPITE THE IMPORTANCE OF NUMEROUS PSYCHOSOCIAL FACTORS, AT ITS CORE, DRUG ADDICTION INVOLVES A BIOLOGICAL PROCESS: the ability of repeated exposure to a drug of abuse to induce changes in a vulnerable brain that drive the compulsive seeking and taking of drugs, and loss of control over drug use, that define a state of addiction. ... A large body of literature has demonstrated that such ΔFosB induction in D1-type NAc neurons increases an animal's sensitivity to drug as well as natural rewards and promotes drug self-administration, presumably through a process of positive reinforcement
- ^ Ruffle JK (November 2014). "Molecular neurobiology of addiction: what's all the (Δ)FosB about?". The American Journal of Drug and Alcohol Abuse. 40 (6): 428–37. doi:10.3109/00952990.2014.933840. PMID 25083822. S2CID 19157711.
ΔFosB is an essential transcription factor implicated in the molecular and behavioral pathways of addiction following repeated drug exposure. The formation of ΔFosB in multiple brain regions, and the molecular pathway leading to the formation of AP-1 complexes is well understood. The establishment of a functional purpose for ΔFosB has allowed further determination as to some of the key aspects of its molecular cascades, involving effectors such as GluR2 (87,88), Cdk5 (93) and NFkB (100). Moreover, many of these molecular changes identified are now directly linked to the structural, physiological and behavioral changes observed following chronic drug exposure (60,95,97,102). New frontiers of research investigating the molecular roles of ΔFosB have been opened by epigenetic studies, and recent advances have illustrated the role of ΔFosB acting on DNA and histones, truly as a molecular switch (34). As a consequence of our improved understanding of ΔFosB in addiction, it is possible to evaluate the addictive potential of current medications (119), as well as use it as a biomarker for assessing the efficacy of therapeutic interventions (121,122,124). Some of these proposed interventions have limitations (125) or are in their infancy (75). However, it is hoped that some of these preliminary findings may lead to innovative treatments, which are much needed in addiction.
- ^ Biliński P, Wojtyła A, Kapka-Skrzypczak L, Chwedorowicz R, Cyranka M, Studziński T (2012). "Epigenetic regulation in drug addiction". Annals of Agricultural and Environmental Medicine. 19 (3): 491–6. PMID 23020045.
For these reasons, ΔFosB is considered a primary and causative transcription factor in creating new neural connections in the reward centre, prefrontal cortex, and other regions of the limbic system. This is reflected in the increased, stable and long-lasting level of sensitivity to cocaine and other drugs, and tendency to relapse even after long periods of abstinence. These newly constructed networks function very efficiently via new pathways as soon as drugs of abuse are further taken
- ^ Olsen CM (December 2011). "Natural rewards, neuroplasticity, and non-drug addictions". Neuropharmacology. 61 (7): 1109–22. doi:10.1016/j.neuropharm.2011.03.010. PMC 3139704. PMID 21459101.
- ^ Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T (January 2011). "Epigenetics in alternative pre-mRNA splicing". Cell. 144 (1): 16–26. doi:10.1016/j.cell.2010.11.056. PMC 3038581. PMID 21215366.
- ^ Strømme JM, Johannessen B, Skotheim RI (2023). "Deviating alternative splicing as a molecular subtype of microsatellite stable colorectal cancer". JCO Clinical Cancer Informatics. 7 (7): e2200159. doi:10.1200/CCI.22.00159. hdl:10852/108838. PMID 36821799.
- ^ Taggart AJ, DeSimone AM, Shih JS, Filloux ME, Fairbrother WG (June 2012). "Large-scale mapping of branchpoints in human pre-mRNA transcripts in vivo". Nature Structural & Molecular Biology. 19 (7): 719–21. doi:10.1038/nsmb.2327. PMC 3465671. PMID 22705790.
- ^ Fairbrother WG, Yeh RF, Sharp PA, Burge CB (August 2002). "Predictive identification of exonic splicing enhancers in human genes". Science. 297 (5583): 1007–13. Bibcode:2002Sci...297.1007F. doi:10.1126/science.1073774. PMID 12114529. S2CID 8689111.
- ^ Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, et al. (December 2004). "Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform". Molecular Cell. 16 (6): 929–41. doi:10.1016/j.molcel.2004.12.004. PMID 15610736.
- ^ a b David CJ, Manley JL (February 2008). "The search for alternative splicing regulators: new approaches offer a path to a splicing code". Genes & Development. 22 (3): 279–85. doi:10.1101/gad.1643108. PMC 2731647. PMID 18245441.
- ^ Watkins KH, Stewart A, Fairbrother W (December 2009). "A rapid high-throughput method for mapping ribonucleoproteins (RNPs) on human pre-mRNA". Journal of Visualized Experiments. 34 (34): 1622. doi:10.3791/1622. PMC 3152247. PMID 19956082.
- ^ Tuerk C, Gold L (August 1990). "Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase". Science. 249 (4968): 505–10. Bibcode:1990Sci...249..505T. doi:10.1126/science.2200121. PMID 2200121.
- ^ Chang B, Levin J, Thompson WA, Fairbrother WG (March 2010). "High-throughput binding analysis determines the binding specificity of ASF/SF2 on alternatively spliced human pre-mRNAs". Combinatorial Chemistry & High Throughput Screening. 13 (3): 242–52. doi:10.2174/138620710790980522. PMC 3427726. PMID 20015017.
- ^ Sommer, Markus J.; Cha, Sooyoung; Varabyou, Ales; Rincon, Natalia; Park, Sukhwan; Minkin, Ilia; Pertea, Mihaela; Steinegger, Martin; Salzberg, Steven L. (2022-12-15). "Structure-guided isoform identification for the human transcriptome". eLife. 11: e82556. doi:10.7554/eLife.82556. PMC 9812405. PMID 36519529.
- ^ Tapial J, Ha KC, Sterne-Weiler T, Gohr A, Braunschweig U, Hermoso-Pulido A, et al. (October 2017). "An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms". Genome Research. 27 (10): 1759–1768. doi:10.1101/gr.220962.117. PMC 5630039. PMID 28855263.
- ^ Louadi Z, Yuan K, Gress A, Tsoy O, Kalinina OV, Baumbach J, et al. (January 2021). "DIGGER: exploring the functional role of alternative splicing in protein interactions". Nucleic Acids Research. 49 (D1): D309–D318. doi:10.1093/nar/gkaa768. PMC 7778957. PMID 32976589.
- ^ Rodriguez JM, Maietta P, Ezkurdia I, Pietrelli A, Wesselink JJ, Lopez G, et al. (January 2013). "APPRIS: annotation of principal and alternative splice isoforms". Nucleic Acids Research. 41 (Database issue): D110-7. doi:10.1093/nar/gks1058. PMC 3531113. PMID 23161672.
External links
[edit]- A General Definition and Nomenclature for Alternative Splicing Events at SciVee
- AStalavista (Alternative Splicing landscape visualization tool), a method for the computationally exhaustive classification of Alternative Splicing Structures Archived 2009-12-11 at the Wayback Machine
- IsoPred: computationally predicted isoform functions
- Stamms-lab.net: Research Group dealing with alternative Splicing issues and mis-splicing in human diseases
- Alternative Splicing of ion channels in the brain, connected to mental and neurological diseases
- BIPASS: Web Services in Alternative Splicing
