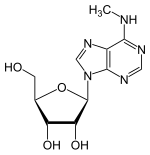
N6-Methyladenosine
| |
| Names | |
|---|---|
| IUPAC name
N6-Methyladenosine
| |
| Systematic IUPAC name
(2R,3S,4R,5R)-2-(Hydroxymethyl)-5-[6-(methylamino)-9H-purin-9-yl]oxolane-2,3-diol | |
| Other names
m6A
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C11H15N5O4 | |
| Molar mass | 281.272 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
N6-Methyladenosine (m6A) was originally identified and partially characterised in the 1970s,[1][2][3][4] and is an abundant modification in mRNA and DNA.[5] It is found within some viruses,[4][3][6][7] and most eukaryotes including mammals,[2][1][8][9] insects,[10] plants[11][12][13] and yeast.[14][15] It is also found in tRNA, rRNA, and small nuclear RNA (snRNA) as well as several long non-coding RNA, such as Xist.[16][17]
The methylation of adenosine is directed by a large m6A methyltransferase complex containing METTL3, which is the subunit that binds S-adenosyl-L-methionine (SAM).[18] In vitro, this methyltransferase complex preferentially methylates RNA oligonucleotides containing GGACU[19] and a similar preference was identified in vivo in mapped m6A sites in Rous sarcoma virus genomic RNA[20] and in bovine prolactin mRNA.[21] More recent studies have characterized other key components of the m6A methyltransferase complex in mammals, including METTL14,[22][23] Wilms tumor 1 associated protein (WTAP),[22][24] VIRMA[25] and METTL5.[26] Following a 2010 speculation of m6A in mRNA being dynamic and reversible,[27] the discovery of the first m6A demethylase, fat mass and obesity-associated protein (FTO) in 2011[28] confirmed this hypothesis and revitalized the interests in the study of m6A. A second m6A demethylase alkB homolog 5 (ALKBH5) was later discovered as well.[29]
The biological functions of m6A are mediated through a group of RNA binding proteins that specifically recognize the methylated adenosine on RNA. These binding proteins are named m6A readers. The YT521-B homology (YTH) domain family of proteins (YTHDF1, YTHDF2, YTHDF3 and YTHDC1) have been characterized as direct m6A readers and have a conserved m6A-binding pocket.[17][30][31][32][33] Insulin-like growth factor-2 mRNA-binding proteins 1, 2, and 3 (IGF2BP1–3) are reported as a novel class of m6A readers.[34] IGF2BPs use K homology (KH) domains to selectively recognize m6A-containing RNAs and promote their translation and stability.[34] These m6A readers, together with m6A methyltransferases (writers) and demethylases (erasers), establish a complex mechanism of m6A regulation in which writers and erasers determine the distributions of m6A on RNA, whereas readers mediate m6A-dependent functions. m6A has also been shown to mediate a structural switch termed m6A switch.[35]
The specificity of m6A installation on mRNA is controlled by exon architecture and exon junction complexes. Exon junction complexes suppress m6A methylation near exon-exon junctions by packaging nearby RNA and protecting it from methylation by the m6A methyltransferase complex. m6A regions in long internal and terminal exons, away from exon-exon junctions and exon junction complexes, escape suppression and can be methylated by the methyltransferase complex. [36]
Species distribution
[edit]Yeast
[edit]In budding yeast (Saccharomyces cerevisiae), the expression of the homologue of METTL3, IME4, is induced in diploid cells in response to nitrogen and fermentable carbon source starvation and is required for mRNA methylation and the initiation of correct meiosis and sporulation.[14][15] mRNAs of IME1 and IME2, key early regulators of meiosis, are known to be targets for methylation, as are transcripts of IME4 itself.[15]
Plants
[edit]In plants, the majority of the m6A is found within 150 nucleotides before the start of the poly(A) tail.[37]
Mutations of MTA, the Arabidopsis thaliana homologue of METTL3, results in embryo arrest at the globular stage. A >90% reduction of m6A levels in mature plants leads to dramatically altered growth patterns and floral homeotic abnormalities.[37]
Mammals
[edit]Mapping of m6A in human and mouse RNA has identified over 18,000 m6A sites in the transcripts of more than 7,000 human genes with a consensus sequence of [G/A/U][G>A]m6AC[U>A/C][16][17][38] consistent with the previously identified motif. The localization of individual m6A sites in many mRNAs is highly similar between human and mouse,[16][17] and transcriptome-wide analysis reveals that m6A is found in regions of high evolutionary conservation.[16] m6A is found within long internal exons and is preferentially enriched within 3' UTRs and around stop codons. m6A within 3' UTRs is also associated with the presence of microRNA binding sites; roughly 2/3 of the mRNAs which contain an m6A site within their 3' UTR also have at least one microRNA binding site.[16] By integrating all m6A sequencing data, a novel database called RMBase has identified and provided ~200,000 sites in the human and mouse genomes corresponding to N6-Methyladenosines (m6A) in RNA.[38]
Precise m6A mapping by m6A-CLIP/IP [39] (briefly m6A-CLIP) revealed that a majority of m6A locates in the last exon of mRNAs in multiple tissues/cultured cells of mouse and human,[39] and the m6A enrichment around stop codons is a coincidence that many stop codons locate round the start of last exons where m6A is truly enriched.[39] The major presence of m6A in last exon (>=70%) allows the potential for 3'UTR regulation, including alternative polyadenylation.[39] The study combining m6A-CLIP with rigorous cell fractionation biochemistry reveals that m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover.[40][41]
m6A is susceptible to dynamic regulation both throughout development and in response to cellular stimuli. Analysis of m6A in mouse brain RNA reveals that m6A levels are low during embryonic development and increase dramatically by adulthood.[16] In mESCs and during mouse development, FTO has been shown to mediated LINE1 RNA m6A demethylation and consequently affect local chromatin state and nearby gene transcription.[42] Additionally, silencing the m6A methyltransferase significantly affects gene expression and alternative RNA splicing patterns, resulting in modulation of the p53 (also known as TP53) signalling pathway and apoptosis.[17]
m6A is also found on the RNA components of R-loops in human and plant cells, where it is involved in regulation of stability of RNA:DNA hybrids. It has been reported to modulate R-loop levels with different outcomes (R-loop resolution and stabilization).[43][44]
The importance of m6A methylation for physiological processes was recently demonstrated. Inhibition of m6A methylation via pharmacological inhibition of cellular methylations or more specifically by siRNA-mediated silencing of the m6A methylase Mettl3 led to the elongation of the circadian period. In contrast, overexpression of Mettl3 led to a shorter period. The mammalian circadian clock, composed of a transcription feedback loop tightly regulated to oscillate with a period of about 24 hours, is therefore extremely sensitive to perturbations in m6A-dependent RNA processing, likely due to the presence of m6A sites within clock gene transcripts.[45][46] The effects of global methylation inhibition on the circadian period in mouse cells can be prevented by ectopic expression of an enzyme from the bacterial methyl metabolism. Mouse cells expressing this bacterial protein were resistant to pharmacological inhibition of methyl metabolism, showing no decrease in mRNA m6A methylation or protein methylation.[47]
Clinical significance
[edit]Considering the versatile functions of m6A in various physiological processes, it is thus not surprising to find links between m6A and numerous human diseases; many originated from mutations or single nucleotide polymorphisms (SNPs) of cognate factors of m6A. The linkages between m6A and numerous cancer types have been indicated in reports that include stomach cancer, prostate cancer, breast cancer, pancreatic cancer, kidney cancer, mesothelioma, sarcoma, and leukaemia.[48][49][50][51][52][53][54][55][56][57][58][59] The impacts of m6A on cancer cell proliferation might be much more profound with more data emerging. The depletion of METTL3 is known to cause apoptosis of cancer cells and reduce invasiveness of cancer cells,[60][61] while the activation of ALKBH5 by hypoxia was shown to cause cancer stem cell enrichment.[62] m6A has also been indicated in the regulation of energy homeostasis and obesity, as FTO is a key regulatory gene for energy metabolism and obesity. SNPs of FTO have been shown to associate with body mass index in human populations and occurrence of obesity and diabetes.[63][64][65][66][67] The influence of FTO on pre-adipocyte differentiation has been suggested.[68][69][70] The connection between m6A and neuronal disorders has also been studied. For instance, neurodegenerative diseases may be affected by m6A as the cognate dopamine signalling was shown to be dependent on FTO and correct m6A methylation on key signalling transcripts.[71] The mutations in HNRNPA2B1, a potential reader of m6A, have been known to cause neurodegeneration.[72] The IGF2BP1–3, a novel class of m6A reader, has oncogenic functions. IGF2BP1–3 knockdown or knockout decreased MYC protein expression, cell proliferation and colony formation in human cancer cell lines.[34] The ZC3H13, a member of the m6A methyltransferase complex, markedly inhibited colorectal cancer cells growth when knocked down.[73]
Additionally, m6A has been reported to impact viral infections. Many RNA viruses including SV40, adenovirus, herpes virus, Rous sarcoma virus, and influenza virus have been known to contain internal m6A methylation on virus genomic RNA.[74] Several more recent studies have revealed that m6A regulators govern the efficiency of infection, replication, translation and transport of RNA viruses such as human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV), and Zika virus (ZIKV).[75][76][77][78][79][80] These results suggest m6A and its cognate factors play crucial roles in regulating virus life cycles and host-viral interactions.
Aside from affecting viruses themselves, m6A modifications can also disrupt the innate immune response. For example, in HBV, m6A modifications were shown to disrupt the recognition of viruses by RIG-1, a pattern recognition receptor in the immune system. Modifications can also disrupt downstream signaling pathways via mechanisms including ubiquitination and changes in the levels of protein expression.[80]
In bacteria
[edit]M6A methylation is also widespread in bacteria, influencing functions such as DNA replication, repair, and gene expression, and prokaryotic defense.
In replication, M6A modifications mark DNA regions where the initiation stage takes place as well as regulates precise timing via the Dam methyltransferase in E. coli.[81][82] Another enzyme, Dam DNA methylase regulates mismatch repair using M6A modifications which influence other repair proteins by recognizing specific mismatches.[83]
In some cases of DNA protection, M6A methylations (along with M4C modifications) play a role in the protection of bacterial DNA by influencing certain endonucleases via the restriction-modification system, decreasing the influence of bacteriophages. One such role is introducing a methyltransferase which recognizes the same target site that restriction enzymes (Type 1 restriction enzymes) attack and modifying it in order to stop such enzymes from attacking bacteria DNA.[84][85]
In Development
[edit]m6A modifications, along with other epigenetic changes, have been shown to play important roles during eukaryotic development. Hematopoietic Stem Cells (HSCs), Neuronal Stem Cells (NSCs) and Primordial Germ Cells (PCGs) have all been shown to undergo m6A modifications during growth and differentiation. Depending on the stage of development, modifications to HSCs can either promote or inhibit stem cell differentiation by affecting the epithelial-to-hemopoietic transition via METTL3 inhibition or depletion. m6A modifications to NSCs can causes changes in brain size, neuron formation, long-term memory, and learning ability. These changes are often caused by inhibition of either METTL or YTHDF readers and writers. In the reproductive system, m6A modifications have been shown to disrupt the maternal-to-zygotic mRNA transition and negatively affect both gamete formation and fertility. Similar to NSCs, inhibition of the METTL and YTHDF families of proteins is often a catalyst for these changes.[86]
References
[edit]- ^ a b Adams JM, Cory S (May 1975). "Modified nucleosides and bizarre 5'-termini in mouse myeloma mRNA". Nature. 255 (5503): 28–33. Bibcode:1975Natur.255...28A. doi:10.1038/255028a0. PMID 1128665. S2CID 4199864.
- ^ a b Desrosiers R, Friderici K, Rottman F (October 1974). "Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells". Proceedings of the National Academy of Sciences of the United States of America. 71 (10): 3971–3975. Bibcode:1974PNAS...71.3971D. doi:10.1073/pnas.71.10.3971. PMC 434308. PMID 4372599.
- ^ a b Aloni Y, Dhar R, Khoury G (October 1979). "Methylation of nuclear simian virus 40 RNAs". Journal of Virology. 32 (1): 52–60. doi:10.1128/JVI.32.1.52-60.1979. PMC 353526. PMID 232187.
- ^ a b Beemon K, Keith J (June 1977). "Localization of N6-methyladenosine in the Rous sarcoma virus genome". Journal of Molecular Biology. 113 (1): 165–179. doi:10.1016/0022-2836(77)90047-X. PMID 196091.
- ^ Ji P, Wang X, Xie N, Li Y (2018). "N6-Methyladenosine in RNA and DNA: An Epitranscriptomic and Epigenetic Player Implicated in Determination of Stem Cell Fate". Stem Cells International. 2018: 3256524. doi:10.1155/2018/3256524. PMC 6199872. PMID 30405719.
- ^ Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR (September 2017). "Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication". Cell Host & Microbe. 22 (3): 377–386.e5. doi:10.1016/j.chom.2017.08.004. PMC 5615858. PMID 28910636.
- ^ Gokhale NS, McIntyre AB, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. (November 2016). "N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection". Cell Host & Microbe. 20 (5): 654–665. doi:10.1016/j.chom.2016.09.015. PMC 5123813. PMID 27773535.
- ^ Wei CM, Gershowitz A, Moss B (January 1976). "5'-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA". Biochemistry. 15 (2): 397–401. doi:10.1021/bi00647a024. PMID 174715.
- ^ Perry RP, Kelley DE, Friderici K, Rottman F (April 1975). "The methylated constituents of L cell messenger RNA: evidence for an unusual cluster at the 5' terminus". Cell. 4 (4): 387–394. doi:10.1016/0092-8674(75)90159-2. PMID 1168101.
- ^ Levis R, Penman S (April 1978). "5'-terminal structures of poly(A)+ cytoplasmic messenger RNA and of poly(A)+ and poly(A)- heterogeneous nuclear RNA of cells of the dipteran Drosophila melanogaster". Journal of Molecular Biology. 120 (4): 487–515. doi:10.1016/0022-2836(78)90350-9. PMID 418182.
- ^ Nichols JL (1979). "In maize poly(A)-containing RNA". Plant Science Letters. 15 (4): 357–361. doi:10.1016/0304-4211(79)90141-X.
- ^ Kennedy TD, Lane BG (June 1979). "Wheat embryo ribonucleates. XIII. Methyl-substituted nucleoside constituents and 5'-terminal dinucleotide sequences in bulk poly(AR)-rich RNA from imbibing wheat embryos". Canadian Journal of Biochemistry. 57 (6): 927–931. doi:10.1139/o79-112. PMID 476526.
- ^ Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG (May 2008). "MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor". The Plant Cell. 20 (5): 1278–1288. doi:10.1105/tpc.108.058883. PMC 2438467. PMID 18505803.
- ^ a b Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA (October 2002). "Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: a potential mechanism for the activity of the IME4 gene". Nucleic Acids Research. 30 (20): 4509–4518. doi:10.1093/nar/gkf573. PMC 137137. PMID 12384598.
- ^ a b c Bodi Z, Button JD, Grierson D, Fray RG (September 2010). "Yeast targets for mRNA methylation". Nucleic Acids Research. 38 (16): 5327–5335. doi:10.1093/nar/gkq266. PMC 2938207. PMID 20421205.
- ^ a b c d e f Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR (June 2012). "Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons". Cell. 149 (7): 1635–1646. doi:10.1016/j.cell.2012.05.003. PMC 3383396. PMID 22608085.
- ^ a b c d e Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. (April 2012). "Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq". Nature. 485 (7397): 201–206. Bibcode:2012Natur.485..201D. doi:10.1038/nature11112. PMID 22575960. S2CID 3517716.
- ^ Bokar JA, Shambaugh ME, Polayes D, Matera AG, Rottman FM (November 1997). "Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase". RNA. 3 (11): 1233–1247. PMC 1369564. PMID 9409616.
- ^ Harper JE, Miceli SM, Roberts RJ, Manley JL (October 1990). "Sequence specificity of the human mRNA N6-adenosine methylase in vitro". Nucleic Acids Research. 18 (19): 5735–5741. doi:10.1093/nar/18.19.5735. PMC 332308. PMID 2216767.
- ^ Kane SE, Beemon K (September 1985). "Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing". Molecular and Cellular Biology. 5 (9): 2298–2306. doi:10.1128/mcb.5.9.2298. PMC 366956. PMID 3016525.
- ^ Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM (September 1984). "Mapping of N6-methyladenosine residues in bovine prolactin mRNA". Proceedings of the National Academy of Sciences of the United States of America. 81 (18): 5667–5671. Bibcode:1984PNAS...81.5667H. doi:10.1073/pnas.81.18.5667. PMC 391771. PMID 6592581.
- ^ a b Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. (February 2014). "A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation". Nature Chemical Biology. 10 (2): 93–95. doi:10.1038/nchembio.1432. PMC 3911877. PMID 24316715.
- ^ Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC (February 2014). "N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells". Nature Cell Biology. 16 (2): 191–198. doi:10.1038/ncb2902. PMC 4640932. PMID 24394384.
- ^ Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. (February 2014). "Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase". Cell Research. 24 (2): 177–189. doi:10.1038/cr.2014.3. PMC 3915904. PMID 24407421.
- ^ Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG, et al. (July 2014). "Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites". Cell Reports. 8 (1): 284–296. doi:10.1016/j.celrep.2014.05.048. PMC 4142486. PMID 24981863.
- ^ van Tran N, Ernst FG, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. (September 2019). "The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112". Nucleic Acids Research. 47 (15): 7719–7733. doi:10.1093/nar/gkz619. PMC 6735865. PMID 31328227.
- ^ He C (December 2010). "Grand challenge commentary: RNA epigenetics?". Nature Chemical Biology. 6 (12): 863–865. doi:10.1038/nchembio.482. PMID 21079590.
- ^ Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. (October 2011). "N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO". Nature Chemical Biology. 7 (12): 885–887. doi:10.1038/nchembio.687. PMC 3218240. PMID 22002720.
- ^ Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. (January 2013). "ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility". Molecular Cell. 49 (1): 18–29. doi:10.1016/j.molcel.2012.10.015. PMC 3646334. PMID 23177736.
- ^ Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. (January 2014). "N6-methyladenosine-dependent regulation of messenger RNA stability". Nature. 505 (7481): 117–120. Bibcode:2014Natur.505..117W. doi:10.1038/nature12730. PMC 3877715. PMID 24284625.
- ^ Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. (June 2015). "N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency". Cell. 161 (6): 1388–1399. doi:10.1016/j.cell.2015.05.014. PMC 4825696. PMID 26046440.
- ^ Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, et al. (November 2014). "Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain". Nature Chemical Biology. 10 (11): 927–929. doi:10.1038/nchembio.1654. PMID 25242552.
- ^ Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. (February 2016). "Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing". Molecular Cell. 61 (4): 507–519. doi:10.1016/j.molcel.2016.01.012. PMID 26876937.
- ^ a b c Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. (March 2018). "Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation". Nature Cell Biology. 20 (3): 285–295. doi:10.1038/s41556-018-0045-z. PMC 5826585. PMID 29476152.
- ^ Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T (February 2015). "N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions". Nature. 518 (7540): 560–564. Bibcode:2015Natur.518..560L. doi:10.1038/nature14234. PMC 4355918. PMID 25719671.
- ^ He PC, Wei J, Dou X, Harada BT, Zhang Z, Ge R, et al. (February 2023). "Exon architecture controls mRNA m6A suppression and gene expression". Science. 379 (6633): 677–682. Bibcode:2023Sci...379..677H. doi:10.1126/science.abj9090. PMC 9990141. PMID 36705538.
- ^ a b Bodi Z, Zhong S, Mehra S, Song J, Graham N, Li H, et al. (2012). "Adenosine Methylation in Arabidopsis mRNA is Associated with the 3' End and Reduced Levels Cause Developmental Defects". Frontiers in Plant Science. 3: 48. doi:10.3389/fpls.2012.00048. PMC 3355605. PMID 22639649.
- ^ a b Sun WJ, Li JH, Liu S, Wu J, Zhou H, Qu LH, Yang JH (January 2016). "RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data". Nucleic Acids Research. 44 (D1): D259–D265. doi:10.1093/nar/gkv1036. PMC 4702777. PMID 26464443.
- ^ a b c d Ke S, Alemu EA, Mertens C, Gantman EC, Fak JJ, Mele A, et al. (October 2015). "A majority of m6A residues are in the last exons, allowing the potential for 3' UTR regulation". Genes & Development. 29 (19): 2037–2053. doi:10.1101/gad.269415.115. PMC 4604345. PMID 26404942.
- ^ Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vågbø CB, Geula S, et al. (May 2017). "m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover". Genes & Development. 31 (10): 990–1006. doi:10.1101/gad.301036.117. PMC 5495127. PMID 28637692.
- ^ Rosa-Mercado NA, Withers JB, Steitz JA (May 2017). "Settling the m6A debate: methylation of mature mRNA is not dynamic but accelerates turnover". Genes & Development. 31 (10): 957–958. doi:10.1101/gad.302695.117. PMC 5495124. PMID 28637691.
- ^ Wei J, Yu X, Yang L, Liu X, Gao B, Huang B, et al. (May 2022). "FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development". Science. 376 (6596): 968–973. Bibcode:2022Sci...376..968W. doi:10.1126/science.abe9582. PMC 9746489. PMID 35511947.
- ^ Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, et al. (January 2020). "N6-methyladenosine regulates the stability of RNA:DNA hybrids in human cells". Nature Genetics. 52 (1): 48–55. doi:10.1038/s41588-019-0549-x. PMC 6974403. PMID 31844323.
- ^ Abakir, A., Ruzov, A. A model for a dual function of N6-methyladenosine in R-loop regulation. Nat Genet (2024). https://doi.org/10.1038/s41588-024-01905-5
- ^ Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. (November 2013). "RNA-methylation-dependent RNA processing controls the speed of the circadian clock". Cell. 155 (4): 793–806. doi:10.1016/j.cell.2013.10.026. PMID 24209618.
- ^ Hastings MH (November 2013). "m(6)A mRNA methylation: a new circadian pacesetter". Cell. 155 (4): 740–741. doi:10.1016/j.cell.2013.10.028. PMID 24209613.
- ^ Fustin JM, Ye S, Rakers C, Kaneko K, Fukumoto K, Yamano M, et al. (May 2020). "Methylation deficiency disrupts biological rhythms from bacteria to humans". Communications Biology. 3 (1): 211. doi:10.1038/s42003-020-0942-0. PMC 7203018. PMID 32376902.
- ^ Akilzhanova A, Nurkina Z, Momynaliev K, Ramanculov E, Zhumadilov Z, Rakhypbekov T, et al. (September 2013). "Genetic profile and determinants of homocysteine levels in Kazakhstan patients with breast cancer". Anticancer Research. 33 (9): 4049–4059. PMID 24023349.
- ^ Reddy SM, Sadim M, Li J, Yi N, Agarwal S, Mantzoros CS, Kaklamani VG (August 2013). "Clinical and genetic predictors of weight gain in patients diagnosed with breast cancer". British Journal of Cancer. 109 (4): 872–881. doi:10.1038/bjc.2013.441. PMC 3749587. PMID 23922112.
- ^ Heiliger KJ, Hess J, Vitagliano D, Salerno P, Braselmann H, Salvatore G, et al. (June 2012). "Novel candidate genes of thyroid tumourigenesis identified in Trk-T1 transgenic mice". Endocrine-Related Cancer. 19 (3): 409–421. doi:10.1530/ERC-11-0387. PMID 22454401.
- ^ Ortega A, Niksic M, Bachi A, Wilm M, Sánchez L, Hastie N, Valcárcel J (January 2003). "Biochemical function of female-lethal (2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing". The Journal of Biological Chemistry. 278 (5): 3040–3047. doi:10.1074/jbc.M210737200. hdl:10261/162976. PMID 12444081.
- ^ Jin DI, Lee SW, Han ME, Kim HJ, Seo SA, Hur GY, et al. (December 2012). "Expression and roles of Wilms' tumor 1-associating protein in glioblastoma". Cancer Science. 103 (12): 2102–2109. doi:10.1111/cas.12022. PMC 7659328. PMID 22957919.
- ^ Lin Y, Ueda J, Yagyu K, Ishii H, Ueno M, Egawa N, et al. (July 2013). "Association between variations in the fat mass and obesity-associated gene and pancreatic cancer risk: a case-control study in Japan". BMC Cancer. 13: 337. doi:10.1186/1471-2407-13-337. PMC 3716552. PMID 23835106.
- ^ Casalegno-Garduño R, Schmitt A, Wang X, Xu X, Schmitt M (October 2010). "Wilms' tumor 1 as a novel target for immunotherapy of leukemia". Transplantation Proceedings. 42 (8): 3309–3311. doi:10.1016/j.transproceed.2010.07.034. PMID 20970678.
- ^ Linnebacher M, Wienck A, Boeck I, Klar E (2010-03-18). "Identification of an MSI-H tumor-specific cytotoxic T cell epitope generated by the (-1) frame of U79260(FTO)". Journal of Biomedicine & Biotechnology. 2010: 841451. doi:10.1155/2010/841451. PMC 2842904. PMID 20339516.
- ^ Machiela MJ, Lindström S, Allen NE, Haiman CA, Albanes D, Barricarte A, et al. (December 2012). "Association of type 2 diabetes susceptibility variants with advanced prostate cancer risk in the Breast and Prostate Cancer Cohort Consortium". American Journal of Epidemiology. 176 (12): 1121–1129. doi:10.1093/aje/kws191. PMC 3571230. PMID 23193118.
- ^ Long J, Zhang B, Signorello LB, Cai Q, Deming-Halverson S, Shrubsole MJ, et al. (2013-04-08). "Evaluating genome-wide association study-identified breast cancer risk variants in African-American women". PLOS ONE. 8 (4): e58350. Bibcode:2013PLoSO...858350L. doi:10.1371/journal.pone.0058350. PMC 3620157. PMID 23593120.
- ^ Kaklamani V, Yi N, Sadim M, Siziopikou K, Zhang K, Xu Y, et al. (April 2011). "The role of the fat mass and obesity associated gene (FTO) in breast cancer risk". BMC Medical Genetics. 12: 52. doi:10.1186/1471-2350-12-52. PMC 3089782. PMID 21489227.
- ^ Pierce BL, Austin MA, Ahsan H (June 2011). "Association study of type 2 diabetes genetic susceptibility variants and risk of pancreatic cancer: an analysis of PanScan-I data". Cancer Causes & Control. 22 (6): 877–883. doi:10.1007/s10552-011-9760-5. PMC 7043136. PMID 21445555.
- ^ Bokar JA (2005-01-01). "The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA". In Grosjean H (ed.). Fine-Tuning of RNA Functions by Modification and Editing. Topics in Current Genetics. Vol. 12. Springer Berlin Heidelberg. pp. 141–177. doi:10.1007/b106365. ISBN 9783540244950.
- ^ Lin S, Choe J, Du P, Triboulet R, Gregory RI (May 2016). "The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells". Molecular Cell. 62 (3): 335–345. doi:10.1016/j.molcel.2016.03.021. PMC 4860043. PMID 27117702.
- ^ Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, et al. (April 2016). "Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA". Proceedings of the National Academy of Sciences of the United States of America. 113 (14): E2047–E2056. Bibcode:2016PNAS..113E2047Z. doi:10.1073/pnas.1602883113. PMC 4833258. PMID 27001847.
- ^ Loos RJ, Yeo GS (January 2014). "The bigger picture of FTO: the first GWAS-identified obesity gene". Nature Reviews. Endocrinology. 10 (1): 51–61. doi:10.1038/nrendo.2013.227. PMC 4188449. PMID 24247219.
- ^ Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. (May 2007). "A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity". Science. 316 (5826): 889–894. Bibcode:2007Sci...316..889F. doi:10.1126/science.1141634. PMC 2646098. PMID 17434869.
- ^ Wang L, Yu Q, Xiong Y, Liu L, Zhang X, Zhang Z, et al. (2013). "Variant rs1421085 in the FTO gene contribute childhood obesity in Chinese children aged 3-6 years". Obesity Research & Clinical Practice. 7 (1): e14–e22. doi:10.1016/j.orcp.2011.12.007. PMID 24331679.
- ^ Kalnina I, Zaharenko L, Vaivade I, Rovite V, Nikitina-Zake L, Peculis R, et al. (September 2013). "Polymorphisms in FTO and near TMEM18 associate with type 2 diabetes and predispose to younger age at diagnosis of diabetes". Gene. 527 (2): 462–468. doi:10.1016/j.gene.2013.06.079. PMID 23860325.
- ^ Karra E, O'Daly OG, Choudhury AI, Yousseif A, Millership S, Neary MT, et al. (August 2013). "A link between FTO, ghrelin, and impaired brain food-cue responsivity". The Journal of Clinical Investigation. 123 (8): 3539–3551. doi:10.1172/jci44403. PMC 3726147. PMID 23867619.
- ^ Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, et al. (December 2014). "FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis". Cell Research. 24 (12): 1403–1419. doi:10.1038/cr.2014.151. PMC 4260349. PMID 25412662.
- ^ Merkestein M, Laber S, McMurray F, Andrew D, Sachse G, Sanderson J, et al. (April 2015). "FTO influences adipogenesis by regulating mitotic clonal expansion". Nature Communications. 6: 6792. Bibcode:2015NatCo...6.6792M. doi:10.1038/ncomms7792. PMC 4410642. PMID 25881961.
- ^ Zhang M, Zhang Y, Ma J, Guo F, Cao Q, Zhang Y, et al. (2015-07-28). "The Demethylase Activity of FTO (Fat Mass and Obesity Associated Protein) Is Required for Preadipocyte Differentiation". PLOS ONE. 10 (7): e0133788. Bibcode:2015PLoSO..1033788Z. doi:10.1371/journal.pone.0133788. PMC 4517749. PMID 26218273.
- ^ Hess ME, Hess S, Meyer KD, Verhagen LA, Koch L, Brönneke HS, et al. (August 2013). "The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry". Nature Neuroscience. 16 (8): 1042–1048. doi:10.1038/nn.3449. PMID 23817550. S2CID 11452560.
- ^ Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. (March 2013). "Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS". Nature. 495 (7442): 467–473. Bibcode:2013Natur.495..467K. doi:10.1038/nature11922. PMC 3756911. PMID 23455423.
- ^ Wang ZL, Li B, Luo YX, Lin Q, Liu SR, Zhang XQ, et al. (January 2018). "Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers". Cell Reports. 22 (1): 286–298. doi:10.1016/j.celrep.2017.12.035. PMID 29298429.
- ^ Narayan P, Rottman FM (1992). "Methylation of mRNA". In Nord FF (ed.). Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology - and Related Areas of Molecular Biology. Vol. 65. pp. 255–285. doi:10.1002/9780470123119.ch7. ISBN 9780470123119. PMID 1315118.
- ^ Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, et al. (May 2016). "Posttranscriptional m(6)A Editing of HIV-1 mRNAs Enhances Viral Gene Expression". Cell Host & Microbe. 19 (5): 675–685. doi:10.1016/j.chom.2016.04.002. PMC 4867121. PMID 27117054.
- ^ Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L (July 2016). "N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression". eLife. 5. doi:10.7554/eLife.15528. PMC 4961459. PMID 27371828.
- ^ Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, et al. (February 2016). "Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells". Nature Microbiology. 1 (4): 16011. doi:10.1038/nmicrobiol.2016.11. PMC 6053355. PMID 27572442.
- ^ Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM (November 2016). "Dynamics of Human and Viral RNA Methylation during Zika Virus Infection". Cell Host & Microbe. 20 (5): 666–673. doi:10.1016/j.chom.2016.10.002. PMC 5155635. PMID 27773536.
- ^ Gokhale NS, McIntyre AB, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. (November 2016). "N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection". Cell Host & Microbe. 20 (5): 654–665. doi:10.1016/j.chom.2016.09.015. PMC 5123813. PMID 27773535.
- ^ a b Moon, Jae-Su; Lee, Wooseong; Cho, Yong-Hee; Kim, Yonghyo; Kim, Geon-Woo (2024-02-28). "The Significance of N6-Methyladenosine RNA Methylation in Regulating the Hepatitis B Virus Life Cycle". Journal of Microbiology and Biotechnology. 34 (2): 233–239. doi:10.4014/jmb.2309.09013. ISSN 1738-8872. PMC 10940779. PMID 37942519.
- ^ O’Brown, Zach Klapholz; Greer, Eric Lieberman (2016), Jeltsch, Albert; Jurkowska, Renata Z. (eds.), "N6-Methyladenine: A Conserved and Dynamic DNA Mark", DNA Methyltransferases - Role and Function, vol. 945, Cham: Springer International Publishing, pp. 213–246, doi:10.1007/978-3-319-43624-1_10, ISBN 978-3-319-43624-1, PMC 5291743, PMID 27826841, retrieved 2024-04-07
- ^ Balzarolo, Melania; Engels, Sander; de Jong, Anja J.; Franke, Katka; van den Berg, Timo K.; Gulen, Muhammet F.; Ablasser, Andrea; Janssen, Edith M.; van Steensel, Bas; Wolkers, Monika C. (March 2021). "m6A methylation potentiates cytosolic dsDNA recognition in a sequence-specific manner". Open Biology. 11 (3). doi:10.1098/rsob.210030. ISSN 2046-2441. PMC 8101014. PMID 33715389.
- ^ Raghunathan, Nalini; Goswami, Sayantan; Leela, Jakku K.; Pandiyan, Apuratha; Gowrishankar, Jayaraman (2019). "A new role for Escherichia coli Dam DNA methylase in prevention of aberrant chromosomal replication". Nucleic Acids Research. 47 (11): 5698–5711. doi:10.1093/nar/gkz242. PMC 6582345. PMID 30957852. Retrieved 2024-04-07.
- ^ Blow, Matthew J.; Clark, Tyson A.; Daum, Chris G.; Deutschbauer, Adam M.; Fomenkov, Alexey; Fries, Roxanne; Froula, Jeff; Kang, Dongwan D.; Malmstrom, Rex R.; Morgan, Richard D.; Posfai, Janos; Singh, Kanwar; Visel, Axel; Wetmore, Kelly; Zhao, Zhiying (2016-02-12). "The Epigenomic Landscape of Prokaryotes". PLOS Genetics. 12 (2): e1005854. doi:10.1371/journal.pgen.1005854. ISSN 1553-7404. PMC 4752239. PMID 26870957.
- ^ Loenen, W. A. M.; Dryden, D. T. F.; Raleigh, E. A.; Wilson, G. G. (2014-01-01). "Type I restriction enzymes and their relatives". Nucleic Acids Research. 42 (1): 20–44. doi:10.1093/nar/gkt847. ISSN 0305-1048. PMC 3874165. PMID 24068554.
- ^ Jiang, Xiulin; Liu, Baiyang; Nie, Zhi; Duan, Lincan; Xiong, Qiuxia; Jin, Zhixian; Yang, Cuiping; Chen, Yongbin (2021-02-21). "The role of m6A modification in the biological functions and diseases". Signal Transduction and Targeted Therapy. 6 (1): 74. doi:10.1038/s41392-020-00450-x. ISSN 2059-3635. PMC 7897327. PMID 33611339.
