Microbial symbiosis and immunity
This article needs attention from an expert in Molecular and Cell Biology or Microbiology. The specific problem is: content unfocused, poor structure, bulk written by inexperienced student editor. (February 2017) |

Long-term close-knit interactions between symbiotic microbes and their host can alter host immune system responses to other microorganisms, including pathogens, and are required to maintain proper homeostasis.[1] The immune system is a host defense system consisting of anatomical physical barriers as well as physiological and cellular responses, which protect the host against harmful microorganisms while limiting host responses to harmless symbionts. Humans are home to 1013 to 1014 bacteria, roughly equivalent to the number of human cells,[2] and while these bacteria can be pathogenic to their host most of them are mutually beneficial to both the host and bacteria.
The human immune system consists of two main types of immunity: innate and adaptive. The innate immune system is made of non-specific defensive mechanisms against foreign cells inside the host including skin as a physical barrier to entry, activation of the complement cascade to identify foreign bacteria and activate necessary cell responses, and white blood cells that remove foreign substances. The adaptive immune system, or acquired immune system, is a pathogen-specific immune response that is carried out by lymphocytes through antigen presentation on MHC molecules to distinguish between self and non-self antigens.
Microbes can promote the development of the host's immune system in the gut and skin, and may help to prevent pathogens from invading. Some release anti-inflammatory products, protecting against parasitic gut microbes. Commensals promote the development of B cells that produce a protective antibody, Immunoglobulin A (IgA). This can neutralize pathogens and exotoxins, and promote the development of immune cells and mucosal immune response. However, microbes have been implicated in human diseases including inflammatory bowel disease, obesity, and cancer.
General principles
[edit]This section needs expansion. You can help by adding to it. (March 2017) |
Microbial symbiosis relies on interspecies communication.[3] between the host and microbial symbionts. Immunity has been historically characterized in multicellular organisms as being controlled by the host immune system, where a perceived foreign substance or cell stimulates an immune response. The end result of this response can vary from clearing of a harmful pathogen to tolerance of a beneficial microbe to an autoimmune response that harms the host itself.
Symbiotic microorganisms have more recently been shown to also be involved in this immune response indicating that the immune response is not isolated to host cells alone. These beneficial microorganisms have been implicated in inhibiting growth of pathogens in the gut and anti-cancer immunity among other responses.
Gastrointestinal tract
[edit]
The human gastrointestinal tract (GI tract) consists of the mouth, pharynx, esophagus, stomach, small intestine, and large intestine, and is a 9-meter-long continuous tube; the largest body surface area exposed to the external environment. The intestine offers nutrients and protection to microbes, enabling them to thrive with an intestinal microbial community of 1014 beneficial and pathogenic bacteria, archaea, viruses, and eukaryotes. In return many of these microbes complete important functions for the host including breakdown of fiber[4] and production of vitamins[5] where gut microbes have at least a role in the production of vitamins such as A, B2, B3, B5, B12, C, D and K.
In the human gut the immune system comes into contact with a large number of foreign microbes, both beneficial and pathogenic. The immune system is capable of protecting the host from these pathogenic microbes without starting unnecessary and harmful immune responses to stimuli. The gastrointestinal microbiota has a direct effect on the human body's immune responses. meaning a regular microbiota is necessary for a healthy host immune system as the body is more susceptible to infectious and non-infectious diseases.
Regulation of immune responses
[edit]Commensal bacteria in the GI tract survive despite the abundance of local immune cells.[6] Homeostasis in the intestine requires stimulation of toll-like receptors by commensal microbes.[6] When mice are raised in germ-free conditions, they lack circulating antibodies, and cannot produce mucus, antimicrobial proteins, or mucosal T-cells.[6] Additionally, mice raised in germ-free conditions lack tolerance and often suffer from hypersensitivity reactions.[6] Maturation of the GI tract is mediated by pattern recognition receptors (PRRs), which recognize non-self pathogen associated molecular patterns (PAMPs) including bacterial cell wall components and nucleic acids.[7] These data suggest that commensal microbes aid in intestinal homeostasis and immune system development.[6]
To prevent constant activation of immune cells and resulting inflammation, hosts and bacteria have evolved to maintain intestinal homeostasis and immune system development.[8] For example, the human symbiont Bacteroides fragilis produces polysaccharide A (PSA), which binds to toll-like receptor 2 (TLR-2) on CD4+ T cells.[9] While TLR2 signaling can activate clearance of peptides, PSA induces an anti-inflammatory response when it binds to TLR2 on CD4+ T cells.[9] Through TLR2 binding, PSA suppresses pro-inflammatory TH17 responses, promoting tolerance and establishing commensal gut colonization.[9]
Commensal gut microbes create a variety of metabolites that bind aryl hydrocarbon receptors (AHR). AHR is a ligand-inducible transcription factor found in immune and epithelial cells and binding of AHR is required for normal immune activation as the lack of AHR binding has been shown to cause over activation of immune cells.[1] These microbial metabolites are crucial for protecting the host from unnecessary inflammation in the gut.
Development of isolated lymphoid tissues
[edit]Microbes trigger development of isolated lymphoid follicles in the small intestine of humans and mice, which are sites of mucosal immune response. Isolated lymphoid follicles (ILFs) collect antigens through M cells, develop germinal centers, and contain many B cells.[10] Gram-negative commensal bacteria trigger the development of inducible lymphoid follicles by releasing peptidogylcans containing diaminopimelic acid during cell division.[10] The peptidoglycans bind to the NOD1 receptor on intestinal epithelial cells.[10] As a result, the intestinal epithelial cells express chemokine ligand 20 (CCL20) and Beta defensin 3.[10] CCL20 and Beta-defensin 3 activate cells which mediate the development of isolated lymphoid tissues, including lymphoid tissue inducer cells and lymphoid tissue organizer cells.[10]
Additionally, there are other mechanisms by which commensals promote maturation of isolated lymphoid follicles. For example, commensal bacteria products bind to TLR2 and TLR4, which results in NF-κB mediated transcription of TNF, which is required for the maturation of mature isolated lymphoid follicles.[11]
Protection against pathogens
[edit]Microbes can prevent growth of harmful pathogens by altering pH, consuming nutrients required for pathogen survival, and secreting toxins and antibodies that inhibit growth of pathogens.[12]
Immunoglobulin A
[edit]IgA prevents entry and colonization of pathogenic bacteria in the gut. It can be found as a monomer, dimer, or tetramer, allowing it to bind multiple antigens simultaneously.[13] IgA coats pathogenic bacterial and viral surfaces (immune exclusion), preventing colonization by blocking their attachment to mucosal cells, and can also neutralize PAMPs.[8][14] IgA promotes the development of TH17 and FOXP3+ regulatory T cells.[15][16] Given its critical function in the GI tract, the number of IgA-secreting plasma cells in the jejunum is greater than the total plasma cell population of the bone marrow, lymph, and spleen combined.[13]
Microbiota-derived signals recruit IgA-secreting plasma cells to mucosal sites.[8] For example, bacteria on the apical surfaces of epithelial cells are phagocytosed by dendritic cells located beneath peyer's patches and in the lamina propria, ultimately leading to differentiation of B cells into plasma cells that secrete IgA specific for intestinal bacteria.[17] The role of microbiota-derived signals in recruiting IgA-secreting plasma cells was confirmed in experiments with antibiotic-treated specific pathogen free and MyD88 KO mice, which have limited commensals and a decreased ability to respond to commensals. The number of intestinal CD11b+ IgA+ plasma cells was reduced in these mice, suggesting the role of commensals in recruiting IgA-secreting plasma cells.[18] Based on this evidence commensal microbes can protect the host from harmful pathogens by stimulating IgA production.
Antimicrobial peptides
[edit]
Members of the microbiota are capable of producing antimicrobial peptides, protecting humans from excessive intestinal inflammation and microbial-associated diseases. Various commensals (primarily Gram-positive bacteria), secrete bacteriocins, peptides which bind to receptors on closely related target cells, forming ion-permeable channels and pores in the cell wall.[19] The resulting efflux of metabolites and cell contents and dissipation of ion gradients causes bacterial cell death.[19] However, bacteriocins can also induce death by translocating into the periplasmic space and cleaving DNA non-specifically (colicin E2), inactivating the ribosome (colicin E3), inhibiting synthesis of peptidoglycan, a major component of the bacterial cell wall (colicin M).[19]
Bacteriocins have immense potential to treat human disease. For example, diarrhea in humans can be caused by a variety of factors, but is often caused by bacteria such as Clostridioides difficile.[19] Microbispora strain ATCC PTA-5024 secretes the bacteriocin microbisporicin, which kills Clostridia by targeting prostaglandin synthesis.[20] Additionally, bacteriocins are particularly promising due to their difference in mechanisms than antibiotics meaning many antibiotic-resistant bacteria are not resistant to these bacteriocins. For example, in vitro growth of methicillin-resistant S. aureus (MRSA) was inhibited by the bacteriocin nisin A, produced by Lactococcus lactis.[19][21] Nisin A inhibits methicillin-resistant S. aureus by binding to the precursor to bacterial cell wall synthesis, lipid II. This hinders the ability to synthesize the cell wall, resulting in increased membrane permeability, disruption of electrochemical gradients, and possible death.[22]
Fortification fucose
[edit]The intestinal epithelium in humans is reinforced with carbohydrates like fucose expressed on the apical surface of epithelial cells.[23] Bacteroides thetaiotaomicron, a bacterial species in the ileum and colon, stimulates the gene encoding fucose, Fut2, in intestinal epithelial cells.[23] In this mutualistic interaction, the intestinal epithelial barrier is fortified and humans are protected against invasion of destructive microbes, while B. thetaiotaomicron benefits because of it can use fucose for energy production and its role in bacterial gene regulation.[23]
Skin
[edit]
The skin microbiota is vital as a line of defense against infection, a physical barrier between the environment and the inside of the host. Commensal microbes that live on the skin, such as Staphylococcus epidermidis, produce antimicrobial peptides (AMPs) that aid the host immune system.[24] These AMPs signal immune responses and maintain an inflammatory homeostasis by modulating the release of cytokines.[24] S. epidermidis secretes a small molecule AMP which leads to increased expression of Human β-defensins.[24] S. epidermidis also stimulates IL-17A+ CD8+ T cells production that increases host immunity.[25]
Exposure to these skin commensal bacteria early in development is crucial for host tolerance of these microbes as T cell encounters allow commensal antigen presentation to be common during development.[26] S. epidermidis and other important microflora work similarly to support homeostasis and general health in areas all over the human body such as the oral cavity, vagina, gastrointestinal tract, and oropharynx.[24]
Role in disease
[edit]An equilibrium of symbionts and pathobionts is critical to fight off outside pathogens and avoid many harmful disorders. Dysbiosis, or imbalances in the bacterial composition of the intestine, has been implicated in inflammatory bowel disease, obesity, and allergic diseases in humans and other animals.[27]
Cancer
[edit]
Gut microbes may play a role in cancer development through a variety of mechanisms. Sulfate-reducing bacteria produce hydrogen sulfide, which results in genomic DNA damage.[28] Higher rates of colon cancer are associated with higher amounts of sulfate-reducing bacteria in the gut.[28] Additionally, anaerobic bacteria in the colon transform primary bile acids into secondary bile acid which has been implicated in colorectal carcinogenesis.[28] Gut bacteria metabolites such as short-chain fatty acids (SCFAs), B vitamins and N1, N12-diacetylspermine have also been implicated in suppressing colorectal cancer.[1] Gram-negative bacteria produce lipopolysaccharide (LPS), which binds to TLR-4 and through TGF-β signaling, leads to the expression of growth factors and inflammatory mediators that promote neoplasia.[28]
Members of a healthy gut microbiome have been shown to increase interferon-γ-producing CD8 T-cells and tumor-infiltrating dendritic cells (TILs) in the intestine.[29] Not only do these CD8 T-cells enhance resistance against intracellular pathogens such as Listeria monocytogenes but they also have been shown to be important in anti-cancer immunity specifically against MC38 adenocarcinoma where they along with the TILs increase MHC I expression.[29]
Allergic and immune disorders
[edit]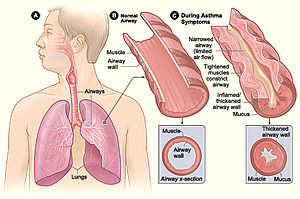
The human microbiome modulates the host immune in positive ways to help defend itself from potential pathogens but can also lead to immune overreactions to foreign substances, even sometimes attacking the host itself. Inflammatory bowel disease (IBD) and asthma are two disorders that have been found to be impacted by microbiota metabolites causing immune reactions. Short-chain fatty acids (SCFAs) have been linked to a decrease in allergic inflammation in asthma[30] while both SCFAs and B vitamins have been shown to decrease IBD inflammation.[31]
SCFAs (acetate, butyrate and propionate) are metabolites created by bacteria in the gut, these molecules then inhibit histone deacetylases (HDACs) as well as G protein-coupled receptors, acting as signaling molecules.[1] Inhibition of HDACs downregulates nuclear factor-κB (NF-κB) and the pro-inflammatory tumor necrosis factor (TNF) as well as having anti-inflammatory effects on macrophages and dendritic cells.[1]
Activation of mucosal immunity and the intestinal microbiota may contribute to inflammatory bowel disease. Many bacteria cause inflammation in the gut including Escherichia coli, which replicate in macrophages and secretes cytokine tumor necrosis factor.[32] However, some bacteria, including the human symbiont B. fragilis, may prevent colitis by producing polysaccharide A (PSA).[33] PSA induces production of IL-10, an immunosuppressive cytokine that suppresses inflammation.[34] Treatment of bone-marrow-derived dendritic cells and naïve CD4+ T cells with purified PSA resulted in increased IL-10 production.[34]
To mimic colitis and activate inflammatory T cells in an experimental condition, wild-type mice were treated with trinitrobenzen sulphonic acid (TNBS).[34] Thereafter, these mice were given PSA orally. Pro-inflammatory cytokine expression (IL-17a and TNFα) in CD4+ cells was measured with ELISA. The researchers found that compared to the CD4+ cells in the control mice, CD4+ cells in PSA-treated mice produced reduced levels of the pro-inflammatory cytokines IL-17a and TNFα.[34] Furthermore, after intestinal colonization with B. fragilis, IL-23 expression by splenocytes was markedly reduced.[34] These data suggest that PSA secreted by B. fragilis suppresses the inflammatory process during colitis by leading to increased production of IL-10 and decreased production of IL-17, TNFα, and IL-23.[34]
Commensal bacteria may also regulate immune responses that cause allergies. For example, commensal bacteria stimulate TLR4, which may inhibit allergic responses to food.[35]
Metabolic disorders
[edit]Major metabolic diseases have been found to be impacted by gut microbiota metabolites, including heart disease, kidney disease, type 2 diabetes and obesity.[1] Breakdown of L-carnitine from red meat by gut microbes into trimethylamine N-oxide (TMAO) has been associated with atherosclerosis, which can lead to obesity, heart disease and type 2 diabetes[36] while both heart and kidney disease events can be predicted by high free p-Cresol levels.[37] SCFAs modulates renin secretion by binding Olfr78, lowering blood pressure and decreasing the risk of kidney disease.[38]
Studies with germ-free mice have suggested that the absence of gut microbes protects against obesity.[39] While the exact mechanism by which microbes play a role in obesity has yet to be elucidated, it has been hypothesized that the intestinal microbiota is involved in converting food to usable energy and fat storage.[39]
Neurological disorders
[edit]Gut microbiota impacts many facets of human health, even neurological disorders that can be caused by molecule or hormone imbalance. Autism spectrum disorder (ASD),[1] central nervous system dysfunction[1] and depression[40] have all been found to be impacted by the microbiota.

While ASD is regularly described by behavioral differences it also can present with gastrointestinal symptoms.[41] Dysbiosis of the GI tract has been noted in some ASD individuals, leading to an increased intestinal permeability.[41] In the mouse model mice with ASD and GI tract dysbiosis (maternal immune activation) increased intestinal permeability was found as was corrected by the introduction of human gut bacterial symbiont B. fragilis.[41]
Microglia development have a pivotal role in central nervous system dysfunction, bacterial metabolite SCFAs regulate microglia homeostasis that is crucial for regular CNS development.[42] Also pivotal for brain development is the creation of tight junctions at the blood-brain barrier in order to control passage between the blood and brain. Germ-free mice have increased blood-brain barrier permeability due to decreased expression of tight junction proteins occludin and claudin-5 as compared to normal gut microbiota mice.[43]
Butyrate-producing bacteria and the dopamine metabolite 3,4-dihydroxyphenylacetic acid have been linked to higher quality of life indicators while γ-aminobutyric acid has been linked to higher levels of depression.[40]
References
[edit]- ^ a b c d e f g h Rooks, Michelle G.; Garrett, Wendy S. (2016-05-27). "Gut microbiota, metabolites and host immunity". Nature Reviews Immunology. 16 (6): 341–352. doi:10.1038/nri.2016.42. ISSN 1474-1733. PMC 5541232. PMID 27231050.
- ^ Mazmanian, Sarkis (2006). "The love–hate relationship between bacterial polysaccharides and the host immune system". Nature Reviews Immunology. 849–858 (11): 849–858. doi:10.1038/nri1956. PMID 17024229. S2CID 20380038.
- ^ McKenney David; Brown Kathryn; Allison David (1995). "Influence of Pseudomonas aeruginosa Exoproducts on Virulence Factor Production in Burkholderia cepacia: Evidence of Interspecies Communication". Journal of Bacteriology. 177 (23): 6989–6991. doi:10.1128/jb.177.23.6989-6992.1995. PMC 177571. PMID 7592496.
- ^ Holscher, Hannah D. (2017-03-04). "Dietary fiber and prebiotics and the gastrointestinal microbiota". Gut Microbes. 8 (2): 172–184. doi:10.1080/19490976.2017.1290756. ISSN 1949-0984. PMC 5390821. PMID 28165863.
- ^ LeBlanc, Jean Guy; Milani, Christian; de Giori, Graciela Savoy; Sesma, Fernando; van Sinderen, Douwe; Ventura, Marco (April 2013). "Bacteria as vitamin suppliers to their host: a gut microbiota perspective". Current Opinion in Biotechnology. 24 (2): 160–168. doi:10.1016/j.copbio.2012.08.005. hdl:11336/2561. ISSN 1879-0429. PMID 22940212.
- ^ a b c d e Brown, E.M. (2013). "A fresh look at the hygiene hypothesis: How intestinal microbial exposure drives immune effector responses in atopic disease". Seminars in Immunology. 25 (5): 378–387. doi:10.1016/j.smim.2013.09.003. PMID 24209708.
- ^ Palm, Noah W.; de Zoete, Marcel R.; Flavell, Richard A. (August 2015). "Immune-microbiota interactions in health and disease". Clinical Immunology. 159 (2): 122–127. doi:10.1016/j.clim.2015.05.014. ISSN 1521-6616. PMC 4943041. PMID 26141651.
- ^ a b c Cerf-Bensussan, Nadine; Gaboriau-Routhiau, Valérie (2010-10-01). "The immune system and the gut microbiota: friends or foes?". Nature Reviews Immunology. 10 (10): 735–744. doi:10.1038/nri2850. PMID 20865020. S2CID 13257259.
- ^ a b c Round, June L.; Lee, S. Melanie; Li, Jennifer; Tran, Gloria; Jabri, Bana; Chatila, Talal A.; Mazmanian, Sarkis K. (2011-05-20). "The Toll-like receptor pathway establishes commensal gut colonization". Science. 332 (6032): 974–977. doi:10.1126/science.1206095. PMC 3164325. PMID 21512004.
- ^ a b c d e Eberl, G.; Lochner, M. (2009-09-09). "The development of intestinal lymphoid tissues at the interface of self and microbiota". Mucosal Immunology. 2 (6): 478–485. doi:10.1038/mi.2009.114. PMID 19741595.
- ^ Bouskra, Djahida; Brézillon, Christophe; Bérard, Marion; Werts, Catherine; Varona, Rosa; Boneca, Ivo Gomperts; Eberl, Gérard (2008-11-27). "Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis". Nature. 456 (7221): 507–510. Bibcode:2008Natur.456..507B. doi:10.1038/nature07450. PMID 18987631. S2CID 205215248.
- ^ Kamada, N (2013). "Control of pathogens and pathobionts by the gut microbiota". Nature Immunology. 14 (7): 685–690. doi:10.1038/ni.2608. PMC 4083503. PMID 23778796.
- ^ a b Kindt, Thomas J.; Goldsby, Richard A.; Osborne, Barbara A.; Kuby, Janis (23 October 2006). Kuby Immunology. W. H. Freeman. pp. 90–92. ISBN 9781429203944.
- ^ Mantis, N. J.; Rol, N.; Corthésy, B. (2011-11-01). "Secretory IgA's complex roles in immunity and mucosal homeostasis in the gut". Mucosal Immunology. 4 (6): 603–611. doi:10.1038/mi.2011.41. PMC 3774538. PMID 21975936.
- ^ Macpherson, AJ (2008). "The immune geography of IgA induction and function". Mucosal Immunology. 1 (1): 11–22. doi:10.1038/mi.2007.6. PMID 19079156.
- ^ Kamada, N (2013). "Role of the gut microbiota in immunity and inflammatory disease". Nature Reviews Immunology. 13 (5): 321–335. doi:10.1038/nri3430. PMID 23618829. S2CID 205491968.
- ^ Hooper Lora V., Bry Lynn, Falk Per G., Gordon Jeffrey I. (1998). "Host–microbial symbiosis in the mammalian intestine: exploring an internal ecosystem". BioEssays. 20 (4): 336–343. doi:10.1002/(sici)1521-1878(199804)20:4<336::aid-bies10>3.3.co;2-j. PMID 9619105.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kunisawa, Jun; Gohda, Masashi; Hashimoto, Eri; Ishikawa, Izumi; Higuchi, Morio; Suzuki, Yuji; Goto, Yoshiyuki; Panea, Casandra; Ivanov, Ivaylo I. (2013-04-23). "Microbe-dependent CD11b+ IgA+ plasma cells mediate robust early-phase intestinal IgA responses in mice". Nature Communications. 4: 1772. Bibcode:2013NatCo...4.1772K. doi:10.1038/ncomms2718. PMC 3644083. PMID 23612313.
- ^ a b c d e Hammami, Riadh; Fernandez, Benoit; Lacroix, Christophe; Fliss, Ismail (2012-10-30). "Anti-infective properties of bacteriocins: an update". Cellular and Molecular Life Sciences. 70 (16): 2947–2967. doi:10.1007/s00018-012-1202-3. PMC 11113238. PMID 23109101. S2CID 16228657.
- ^ Castiglione, Franca; Lazzarini, Ameriga; Carrano, Lucia; Corti, Emiliana; Ciciliato, Ismaela; Gastaldo, Luciano; Candiani, Paolo; Losi, Daniele; Marinelli, Flavia (2008-01-25). "Determining the Structure and Mode of Action of Microbisporicin, a Potent Lantibiotic Active Against Multiresistant Pathogens". Chemistry & Biology. 15 (1): 22–31. doi:10.1016/j.chembiol.2007.11.009. PMID 18215770.
- ^ Piper, C.; Draper, L. A.; Cotter, P. D.; Ross, R. P.; Hill, C. (2009-09-01). "A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species". Journal of Antimicrobial Chemotherapy. 64 (3): 546–551. doi:10.1093/jac/dkp221. PMID 19561147.
- ^ Hsu, Shang-Te D.; Breukink, Eefjan; Tischenko, Eugene; Lutters, Mandy A. G.; de Kruijff, Ben; Kaptein, Robert; Bonvin, Alexandre M. J. J.; van Nuland, Nico A. J. (2004-10-01). "The nisin–lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics". Nature Structural & Molecular Biology. 11 (10): 963–967. doi:10.1038/nsmb830. hdl:1874/385449. PMID 15361862. S2CID 13181577.
- ^ a b c Goto, Yoshiyuki; Kiyono, Hiroshi (2012). "Epithelial barrier: an interface for the cross-communication between gut flora and immune system". Immunological Reviews. 245 (1): 147–163. doi:10.1111/j.1600-065X.2011.01078.x. PMID 22168418. S2CID 22187069.
- ^ a b c d Gallo Richard L., Nakatsuji Teruaki (2011). "Microbial symbiosis with the innate immune defense system of the skin". Journal of Investigative Dermatology. 131 (10): 1974–1980. doi:10.1038/jid.2011.182. PMC 3174284. PMID 21697881.
- ^ Naik, Shruti; Bouladoux, Nicolas; Linehan, Jonathan L.; Han, Seong-Ji; Harrison, Oliver J.; Wilhelm, Christoph; Conlan, Sean; Himmelfarb, Sarah; Byrd, Allyson L.; Deming, Clayton; Quinones, Mariam (2015-04-02). "Commensal–dendritic-cell interaction specifies a unique protective skin immune signature". Nature. 520 (7545): 104–108. Bibcode:2015Natur.520..104N. doi:10.1038/nature14052. ISSN 0028-0836. PMC 4667810. PMID 25539086.
- ^ Scharschmidt, Tiffany C. (January 2017). "Establishing tolerance to commensal skin bacteria: timing is everything". Dermatologic Clinics. 35 (1): 1–9. doi:10.1016/j.det.2016.07.007. ISSN 0733-8635. PMC 5130113. PMID 27890233.
- ^ DeGruttola, Arianna K.; Low, Daren; Mizoguchi, Atsushi; Mizoguchi, Emiko (2017-02-25). "Current understanding of dysbiosis in disease in human and animal models". Inflammatory Bowel Diseases. 22 (5): 1137–1150. doi:10.1097/MIB.0000000000000750. PMC 4838534. PMID 27070911.
- ^ a b c d Hullar, Meredith A. J.; Burnett-Hartman, Andrea N.; Lampe, Johanna W. (2014-01-01). "Gut Microbes, Diet, and Cancer". Advances in Nutrition and Cancer. Cancer Treatment and Research. Vol. 159. pp. 377–399. doi:10.1007/978-3-642-38007-5_22. ISBN 978-3-642-38006-8. ISSN 0927-3042. PMC 4121395. PMID 24114492.
- ^ a b Tanoue, Takeshi; Morita, Satoru; Plichta, Damian R.; Skelly, Ashwin N.; Suda, Wataru; Sugiura, Yuki; Narushima, Seiko; Vlamakis, Hera; Motoo, Iori; Sugita, Kayoko; Shiota, Atsushi (January 2019). "A defined commensal consortium elicits CD8 T cells and anti-cancer immunity". Nature. 565 (7741): 600–605. Bibcode:2019Natur.565..600T. doi:10.1038/s41586-019-0878-z. ISSN 1476-4687. PMID 30675064. S2CID 59159425.
- ^ Min, Booki (2014-02-18). "Faculty of 1000 evaluation for Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis". doi:10.3410/f.718228193.793491060.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Singh, Nagendra; Gurav, Ashish; Sivaprakasam, Sathish; Brady, Evan; Padia, Ravi; Shi, Huidong; Thangaraju, Muthusamy; Prasad, Puttur D.; Manicassamy, Santhakumar; Munn, David H.; Lee, Jeffrey R. (January 2014). "Activation of Gpr109a, Receptor for Niacin and the Commensal Metabolite Butyrate, Suppresses Colonic Inflammation and Carcinogenesis". Immunity. 40 (1): 128–139. doi:10.1016/j.immuni.2013.12.007. ISSN 1074-7613. PMC 4305274. PMID 24412617.
- ^ Sartor, R. Balfour; Mazmanian, Sarkis K. (2012-07-01). "Intestinal Microbes in Inflammatory Bowel Diseases". The American Journal of Gastroenterology Supplements. 1 (1): 15–21. doi:10.1038/ajgsup.2012.4.
- ^ Round, June L.; Mazmanian, Sarkis K. (2017-02-16). "The gut microbiome shapes intestinal immune responses during health and disease". Nature Reviews Immunology. 9 (5): 313–323. doi:10.1038/nri2515. PMC 4095778. PMID 19343057.
- ^ a b c d e f Mazmanian, Sarkis K.; Round, June L.; Kasper, Dennis L. (2008). "A microbial symbiosis factor prevents intestinal inflammatory disease". Nature. 453 (7195): 620–625. Bibcode:2008Natur.453..620M. doi:10.1038/nature07008. PMID 18509436. S2CID 205213521.
- ^ Bashir, Mohamed Elfatih H.; Louie, Steve; Shi, Hai Ning; Nagler-Anderson, Cathryn (2004-06-01). "Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy". Journal of Immunology. 172 (11): 6978–6987. doi:10.4049/jimmunol.172.11.6978. PMID 15153518.
- ^ Terry, Paul; Chen, Jiangang (2013-04-11). "Faculty of 1000 evaluation for Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis". doi:10.3410/f.717998892.793474469.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Meijers, Björn K.I.; Claes, Kathleen; Bammens, Bert; de Loor, Henriette; Viaene, Liesbeth; Verbeke, Kristin; Kuypers, Dirk; Vanrenterghem, Yves; Evenepoel, Pieter (July 2010). "p-Cresol and Cardiovascular Risk in Mild-to-Moderate Kidney Disease". Clinical Journal of the American Society of Nephrology. 5 (7): 1182–1189. doi:10.2215/CJN.07971109. ISSN 1555-9041. PMC 2893077. PMID 20430946.
- ^ Persson, A Erik G; Carlström, Mattias (2013-05-21). "Faculty of 1000 evaluation for Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation". doi:10.3410/f.717978217.793476920.
{{cite journal}}: Cite journal requires|journal=(help) - ^ a b Carding, Simon; Verbeke, Kristin; Vipond, Daniel T.; Corfe, Bernard M.; Owen, Lauren J. (2015-02-02). "Dysbiosis of the gut microbiota in disease". Microbial Ecology in Health and Disease. 26: 26191. doi:10.3402/mehd.v26.26191. ISSN 0891-060X. PMC 4315779. PMID 25651997.
- ^ a b Valles-Colomer, Mireia; Falony, Gwen; Darzi, Youssef; Tigchelaar, Ettje F.; Wang, Jun; Tito, Raul Y.; Schiweck, Carmen; Kurilshikov, Alexander; Joossens, Marie; Wijmenga, Cisca; Claes, Stephan (April 2019). "The neuroactive potential of the human gut microbiota in quality of life and depression" (PDF). Nature Microbiology. 4 (4): 623–632. doi:10.1038/s41564-018-0337-x. ISSN 2058-5276. PMID 30718848. S2CID 59603019.
- ^ a b c Hsiao, Elaine Y.; McBride, Sara W.; Hsien, Sophia; Sharon, Gil; Hyde, Embriette R.; McCue, Tyler; Codelli, Julian A.; Chow, Janet; Reisman, Sarah E.; Petrosino, Joseph F.; Patterson, Paul H. (2013-12-19). "The microbiota modulates gut physiology and behavioral abnormalities associated with autism". Cell. 155 (7): 1451–1463. doi:10.1016/j.cell.2013.11.024. ISSN 0092-8674. PMC 3897394. PMID 24315484.
- ^ Davidovic, Laetitia (2015-06-22). "Faculty of 1000 evaluation for Host microbiota constantly control maturation and function of microglia in the CNS". doi:10.3410/f.725528105.793507643.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Braniste, Viorica; Al-Asmakh, Maha; Kowal, Czeslawa; Anuar, Farhana; Abbaspour, Afrouz; Tóth, Miklós; Korecka, Agata; Bakocevic, Nadja; Ng, Lai Guan; Kundu, Parag; Gulyás, Balázs (2014-11-19). "The gut microbiota influences blood-brain barrier permeability in mice". Science Translational Medicine. 6 (263): 263ra158. doi:10.1126/scitranslmed.3009759. ISSN 1946-6234. PMC 4396848. PMID 25411471.
