Genotype
| Part of a series on |
| Biology |
|---|
The genotype of an organism is its complete set of genetic material.[1] Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location.[2] The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous.
Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism.[3] The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plant.[4] However, other traits are only partially influenced by genotype. These traits are often called complex traits because they are influenced by additional factors, such as environmental and epigenetic factors. Not all individuals with the same genotype look or act the same way because appearance and behavior are modified by environmental and growing conditions. Likewise, not all organisms that look alike necessarily have the same genotype.
The term genotype was coined by the Danish botanist Wilhelm Johannsen in 1903.[5]
Phenotype
[edit]Any given gene will usually cause an observable change in an organism, known as the phenotype. The terms genotype and phenotype are distinct for at least two reasons:
- To distinguish the source of an observer's knowledge (one can know about genotype by observing DNA; one can know about phenotype by observing outward appearance of an organism).
- Genotype and phenotype are not always directly correlated. Some genes only express a given phenotype in certain environmental conditions. Conversely, some phenotypes could be the result of multiple genotypes. The genotype is commonly mixed up with the phenotype which describes the result of both the genetic and the environmental factors giving the observed expression (e.g. blue eyes, hair color, or various hereditary diseases).
A simple example to illustrate genotype as distinct from phenotype is the flower colour in pea plants (see Gregor Mendel). There are three available genotypes, PP (homozygous dominant), Pp (heterozygous), and pp (homozygous recessive). All three have different genotypes but the first two have the same phenotype (purple) as distinct from the third (white).
A more technical example to illustrate genotype is the single-nucleotide polymorphism or SNP. A SNP occurs when corresponding sequences of DNA from different individuals differ at one DNA base, for example where the sequence AAGCCTA changes to AAGCTTA.[6] This contains two alleles : C and T. SNPs typically have three genotypes, denoted generically AA Aa and aa. In the example above, the three genotypes would be CC, CT and TT. Other types of genetic marker, such as microsatellites, can have more than two alleles, and thus many different genotypes.
Penetrance is the proportion of individuals showing a specified genotype in their phenotype under a given set of environmental conditions.[7]
Mendelian inheritance
[edit]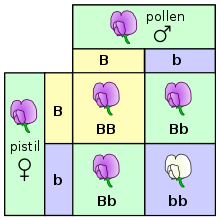
Traits that are determined exclusively by genotype are typically inherited in a Mendelian pattern. These laws of inheritance were described extensively by Gregor Mendel, who performed experiments with pea plants to determine how traits were passed on from generation to generation.[8] He studied phenotypes that were easily observed, such as plant height, petal color, or seed shape.[8] He was able to observe that if he crossed two true-breeding plants with distinct phenotypes, all the offspring would have the same phenotype. For example, when he crossed a tall plant with a short plant, all the resulting plants would be tall. However, when he self-fertilized the plants that resulted, about 1/4 of the second generation would be short. He concluded that some traits were dominant, such as tall height, and others were recessive, like short height. Though Mendel was not aware at the time, each phenotype he studied was controlled by a single gene with two alleles. In the case of plant height, one allele caused the plants to be tall, and the other caused plants to be short. When the tall allele was present, the plant would be tall, even if the plant was heterozygous. In order for the plant to be short, it had to be homozygous for the recessive allele.[8][9]
One way this can be illustrated is using a Punnett square. In a Punnett square, the genotypes of the parents are placed on the outside. An uppercase letter is typically used to represent the dominant allele, and a lowercase letter is used to represent the recessive allele. The possible genotypes of the offspring can then be determined by combining the parent genotypes.[10] In the example on the right, both parents are heterozygous, with a genotype of Bb. The offspring can inherit a dominant allele from each parent, making them homozygous with a genotype of BB. The offspring can inherit a dominant allele from one parent and a recessive allele from the other parent, making them heterozygous with a genotype of Bb. Finally, the offspring could inherit a recessive allele from each parent, making them homozygous with a genotype of bb. Plants with the BB and Bb genotypes will look the same, since the B allele is dominant. The plant with the bb genotype will have the recessive trait.
These inheritance patterns can also be applied to hereditary diseases or conditions in humans or animals.[11][12][13] Some conditions are inherited in an autosomal dominant pattern, meaning individuals with the condition typically have an affected parent as well. A classic pedigree for an autosomal dominant condition shows affected individuals in every generation.[11][12][13]

Other conditions are inherited in an autosomal recessive pattern, where affected individuals do not typically have an affected parent. Since each parent must have a copy of the recessive allele in order to have an affected offspring, the parents are referred to as carriers of the condition.[11][12][13] In autosomal conditions, the sex of the offspring does not play a role in their risk of being affected. In sex-linked conditions, the sex of the offspring affects their chances of having the condition. In humans, females inherit two X chromosomes, one from each parent, while males inherit an X chromosome from their mother and a Y chromosome from their father. X-linked dominant conditions can be distinguished from autosomal dominant conditions in pedigrees by the lack of transmission from fathers to sons, since affected fathers only pass their X chromosome to their daughters.[13][11][14] In X-linked recessive conditions, males are typically affected more commonly because they are hemizygous, with only one X chromosome. In females, the presence of a second X chromosome will prevent the condition from appearing. Females are therefore carriers of the condition and can pass the trait on to their sons.[13][11][14]
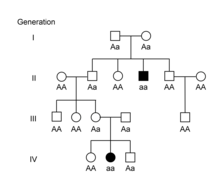
Mendelian patterns of inheritance can be complicated by additional factors. Some diseases show incomplete penetrance, meaning not all individuals with the disease-causing allele develop signs or symptoms of the disease.[13][15][16] Penetrance can also be age-dependent, meaning signs or symptoms of disease are not visible until later in life. For example, Huntington disease is an autosomal dominant condition, but up to 25% of individuals with the affected genotype will not develop symptoms until after age 50.[17] Another factor that can complicate Mendelian inheritance patterns is variable expressivity, in which individuals with the same genotype show different signs or symptoms of disease.[13][15][16] For example, individuals with polydactyly can have a variable number of extra digits.[15][16]
Non-Mendelian inheritance
[edit]Many traits are not inherited in a Mendelian fashion, but have more complex patterns of inheritance.
Incomplete dominance
[edit]For some traits, neither allele is completely dominant. Heterozygotes often have an appearance somewhere in between those of homozygotes.[18][19] For example, a cross between true-breeding red and white Mirabilis jalapa results in pink flowers.[19]
Codominance
[edit]Codominance refers to traits in which both alleles are expressed in the offspring in approximately equal amounts.[20] A classic example is the ABO blood group system in humans, where both the A and B alleles are expressed when they are present. Individuals with the AB genotype have both A and B proteins expressed on their red blood cells.[20][18]
Epistasis
[edit]Epistasis is when the phenotype of one gene is affected by one or more other genes.[21] This is often through some sort of masking effect of one gene on the other.[22] For example, the "A" gene codes for hair color, a dominant "A" allele codes for brown hair, and a recessive "a" allele codes for blonde hair, but a separate "B" gene controls hair growth, and a recessive "b" allele causes baldness. If the individual has the BB or Bb genotype, then they produce hair and the hair color phenotype can be observed, but if the individual has a bb genotype, then the person is bald which masks the A gene entirely.
Polygenic traits
[edit]A polygenic trait is one whose phenotype is dependent on the additive effects of multiple genes. The contributions of each of these genes are typically small and add up to a final phenotype with a large amount of variation. A well studied example of this is the number of sensory bristles on a fly.[23] These types of additive effects is also the explanation for the amount of variation in human eye color.
Genotyping
[edit]Genotyping refers to the method used to determine an individual's genotype. There are a variety of techniques that can be used to assess genotype. The genotyping method typically depends on what information is being sought. Many techniques initially require amplification of the DNA sample, which is commonly done using PCR.
Some techniques are designed to investigate specific SNPs or alleles in a particular gene or set of genes, such as whether an individual is a carrier for a particular condition. This can be done via a variety of techniques, including allele specific oligonucleotide (ASO) probes or DNA sequencing.[24][25] Tools such as multiplex ligation-dependent probe amplification can also be used to look for duplications or deletions of genes or gene sections.[25] Other techniques are meant to assess a large number of SNPs across the genome, such as SNP arrays.[24][25] This type of technology is commonly used for genome-wide association studies.
Large-scale techniques to assess the entire genome are also available. This includes karyotyping to determine the number of chromosomes an individual has and chromosomal microarrays to assess for large duplications or deletions in the chromosome.[24][25] More detailed information can be determined using exome sequencing, which provides the specific sequence of all DNA in the coding region of the genome, or whole genome sequencing, which sequences the entire genome including non-coding regions.[24][25]
Genotype encoding
[edit]In linear models, the genotypes can be encoded in different manners. Let us consider a biallelic locus with two possible alleles, encoded by and . We consider to correspond to the dominant allele to the reference allele . The following table details the different encoding.[26]
| Genotype | |||
|---|---|---|---|
| Additive encoding | 0 | 1 | 2 |
| Dominant encoding | 0 | 1 | 1 |
| Recessive encoding | 0 | 0 | 1 |
| Codominant encoding | 0,0 | 0,1 | 1,0 |
See also
[edit]References
[edit]- ^ "What is genotype? What is phenotype? – pgEd". pged.org. Retrieved 2020-06-22.
- ^ "Genotype". Genome.gov. Retrieved 2021-11-09.
- ^ Pierce, Benjamin (2020). Genetics A Conceptual Approach. NY, New York: Macmillian. ISBN 978-1-319-29714-5.
- ^ Alberts B, Bray D, Hopkin K, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2014). Essential Cell Biology (4th ed.). New York, NY: Garland Science. p. 659. ISBN 978-0-8153-4454-4.
- ^ Johannsen W (1903). "Om arvelighed i samfund og i rene linier". Oversigt Birdy over Det Kongelige Danske Videnskabernes Selskabs Forhandlingerm (in Danish). 3: 247–70. German ed. "Erblichkeit in Populationen und in reinen Linien" (in German). Jena: Gustav Fischer. 1903. Archived from the original on 2009-05-30. Retrieved 2017-07-19.. Also see his monograph Johannsen W (1905). Arvelighedslærens elementer horse [The Elements of Heredity] (in Danish). Copenhagen.
{{cite book}}: CS1 maint: location missing publisher (link) which was rewritten, enlarged and translated into French as Johannsen W (1905). Elemente der exakten Erblichkeitslehre (in German). Jena: Gustav Fischer. Archived from the original on 2009-05-30. Retrieved 2017-07-19. - ^ Vallente, R. U., PhD. (2020). Single Nucleotide Polymorphism. Salem Press Encyclopedia of Science.
- ^ Allaby, Michael, ed. (2009). A dictionary of zoology (3rd ed.). Oxford: Oxford University Press. ISBN 9780199233410. OCLC 260204631.
- ^ a b c "Gregor Mendel and the Principles of Inheritance | Learn Science at Scitable". www.nature.com. Retrieved 2021-11-15.
- ^ "12.1 Mendel's Experiments and the Laws of Probability – Biology | OpenStax". openstax.org. Retrieved 2021-11-15.
- ^ "3.6: Punnett Squares". Biology LibreTexts. 2016-09-21. Retrieved 2021-11-15.
- ^ a b c d e Alliance, Genetic; Health, District of Columbia Department of (2010-02-17). Classic Mendelian Genetics (Patterns of Inheritance). Genetic Alliance.
- ^ a b c "Mendelian Inheritance". Genome.gov. Retrieved 2021-11-15.
- ^ a b c d e f g Strachan, T. (2018). Human molecular genetics. Andrew P. Read (5th ed.). New York: Garland Science. ISBN 978-0-429-82747-1. OCLC 1083018958.
- ^ a b "4.4.1: Inheritance patterns for X-linked and Y-linked genes". Biology LibreTexts. 2020-06-24. Retrieved 2021-11-15.
- ^ a b c "14.2: Penetrance and Expressivity". Biology LibreTexts. 2021-01-13. Retrieved 2021-11-19.
- ^ a b c "Phenotype Variability: Penetrance and Expressivity | Learn Science at Scitable". www.nature.com. Retrieved 2021-11-19.
- ^ Caron, Nicholas S.; Wright, Galen EB; Hayden, Michael R. (1993), Adam, Margaret P.; Ardinger, Holly H.; Pagon, Roberta A.; Wallace, Stephanie E. (eds.), "Huntington Disease", GeneReviews®, Seattle (WA): University of Washington, Seattle, PMID 20301482, retrieved 2021-11-19
- ^ a b "Genetic Dominance: Genotype-Phenotype Relationships | Learn Science at Scitable". www.nature.com. Retrieved 2021-11-15.
- ^ a b Frizzell, M.A. (2013), "Incomplete Dominance", Brenner's Encyclopedia of Genetics, Elsevier, pp. 58–60, doi:10.1016/b978-0-12-374984-0.00784-1, ISBN 978-0-08-096156-9, retrieved 2021-11-15
- ^ a b Xia, X. (2013), "Codominance", Brenner's Encyclopedia of Genetics, Elsevier, pp. 63–64, doi:10.1016/b978-0-12-374984-0.00278-3, ISBN 978-0-08-096156-9, retrieved 2021-11-15
- ^ Gros, Pierre-Alexis; Nagard, Hervé Le; Tenaillon, Olivier (2009-05-01). "The Evolution of Epistasis and Its Links With Genetic Robustness, Complexity and Drift in a Phenotypic Model of Adaptation". Genetics. 182 (1): 277–293. doi:10.1534/genetics.108.099127. ISSN 0016-6731. PMC 2674823. PMID 19279327.
- ^ Rieger, Rigomar. (1976). Glossary of genetics and cytogenetics : classical and molecular. Michaelis, Arnd,, Green, Melvin M. (4th completely rev. ed.). Berlin: Springer-Verlag. ISBN 0-387-07668-9. OCLC 2202589.
- ^ Mackay, T. F. (December 1995). "The genetic basis of quantitative variation: numbers of sensory bristles of Drosophila melanogaster as a model system". Trends in Genetics. 11 (12): 464–470. doi:10.1016/s0168-9525(00)89154-4. ISSN 0168-9525. PMID 8533161.
- ^ a b c d Jain, Kewal K. (2015), Jain, Kewal K. (ed.), "Molecular Diagnostics in Personalized Medicine", Textbook of Personalized Medicine, New York, NY: Springer, pp. 35–89, doi:10.1007/978-1-4939-2553-7_2, ISBN 978-1-4939-2553-7, retrieved 2021-11-19
- ^ a b c d e Wallace, Stephanie E.; Bean, Lora JH (2020-06-18). Educational Materials – Genetic Testing: Current Approaches. University of Washington, Seattle.
- ^ Schönbach, Christian; Ranganathan, Shoba; Nakai, Kenta, eds. (2018). Encyclopedia of Bioinformatics and Computational Biology. Elsevier Science. p. 174. ISBN 9780128114322.





