V(D)J recombination
V(D)J recombination (variable–diversity–joining rearrangement) is the mechanism of somatic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. It results in the highly diverse repertoire of antibodies/immunoglobulins and T cell receptors (TCRs) found in B cells and T cells, respectively. The process is a defining feature of the adaptive immune system.
V(D)J recombination in mammals occurs in the primary lymphoid organs (bone marrow for B cells and thymus for T cells) and in a nearly random fashion rearranges variable (V), joining (J), and in some cases, diversity (D) gene segments. The process ultimately results in novel amino acid sequences in the antigen-binding regions of immunoglobulins and TCRs that allow for the recognition of antigens from nearly all pathogens including bacteria, viruses, parasites, and worms as well as "altered self cells" as seen in cancer. The recognition can also be allergic in nature (e.g. to pollen or other allergens) or may match host tissues and lead to autoimmunity.
In 1987, Susumu Tonegawa was awarded the Nobel Prize in Physiology or Medicine "for his discovery of the genetic principle for generation of antibody diversity".[1]
Background
[edit]Human antibody molecules (including B cell receptors) are composed of heavy and light chains, each of which contains both constant (C) and variable (V) regions, genetically encoded on three loci:
- The immunoglobulin heavy locus (IGH@) on chromosome 14, containing the gene segments for the immunoglobulin heavy chain.
- The immunoglobulin kappa (κ) locus (IGK@) on chromosome 2, containing the gene segments for one type (κ) of immunoglobulin light chain.
- The immunoglobulin lambda (λ) locus (IGL@) on chromosome 22, containing the gene segments for another type (λ) of immunoglobulin light chain.
Each heavy chain or light chain gene contains multiple copies of three different types of gene segments for the variable regions of the antibody proteins. For example, the human immunoglobulin heavy chain region contains 2 Constant (Cμ and Cδ) gene segments and 44 Variable (V) gene segments, plus 27 Diversity (D) gene segments and 6 Joining (J) gene segments.[2] The light chain genes possess either a single (Cκ) or four (Cλ) Constant gene segments with numerous V and J gene segments but do not have D gene segments.[3] DNA rearrangement causes one copy of each type of gene segment to go in any given lymphocyte, generating an enormous antibody repertoire; roughly 3×1011 combinations are possible, although some are removed due to self reactivity.
Most T cell receptors are composed of a variable alpha chain and a beta chain. The T cell receptor genes are similar to immunoglobulin genes in that they too contain multiple V, D, and J gene segments in their beta chains (and V and J gene segments in their alpha chains) that are rearranged during the development of the lymphocyte to provide that cell with a unique antigen receptor. The T cell receptor in this sense is the topological equivalent to an antigen-binding fragment of the antibody, both being part of the immunoglobulin superfamily.
An autoimmune response is prevented by eliminating cells that self-react. This occurs in the thymus by testing the cell against an array of self antigens expressed through the function of the autoimmune regulator (AIRE). The immunoglobulin lambda light chain locus contains protein-coding genes that can be lost with its rearrangement. This is based on a physiological mechanism and is not pathogenetic for leukemias or lymphomas. A cell persists if it creates a successful product that does not self-react, otherwise it is pruned via apoptosis.
Immunoglobulins
[edit]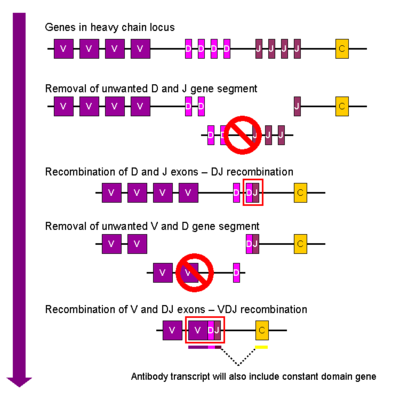
Heavy chain
[edit]In the developing B cell, the first recombination event to occur is between one D and one J gene segment of the heavy chain locus. Any DNA between these two gene segments is deleted. This D-J recombination is followed by the joining of one V gene segment, from a region upstream of the newly formed DJ complex, forming a rearranged VDJ gene segment. All other gene segments between V and D segments are now deleted from the cell's genome. Primary transcript (unspliced RNA) is generated containing the VDJ region of the heavy chain and both the constant mu and delta chains (Cμ and Cδ). (i.e. the primary transcript contains the segments: V-D-J-Cμ-Cδ). The primary RNA is processed to add a polyadenylated (poly-A) tail after the Cμ chain and to remove sequence between the VDJ segment and this constant gene segment. Translation of this mRNA leads to the production of the IgM heavy chain protein.
Light chain
[edit]The kappa (κ) and lambda (λ) chains of the immunoglobulin light chain loci rearrange in a very similar way, except that the light chains lack a D segment. In other words, the first step of recombination for the light chains involves the joining of the V and J chains to give a VJ complex before the addition of the constant chain gene during primary transcription. Translation of the spliced mRNA for either the kappa or lambda chains results in formation of the Ig κ or Ig λ light chain protein.
Assembly of the Ig μ heavy chain and one of the light chains results in the formation of membrane bound form of the immunoglobulin IgM that is expressed on the surface of the immature B cell.
T cell receptors
[edit]During thymocyte development, the T cell receptor (TCR) chains undergo essentially the same sequence of ordered recombination events as that described for immunoglobulins. D-to-J recombination occurs first in the β-chain of the TCR. This process can involve either the joining of the Dβ1 gene segment to one of six Jβ1 segments or the joining of the Dβ2 gene segment to one of six Jβ2 segments.[3] DJ recombination is followed (as above) with Vβ-to-DβJβ rearrangements. All gene segments between the Vβ-Dβ-Jβ gene segments in the newly formed complex are deleted and the primary transcript is synthesized that incorporates the constant domain gene (Vβ-Dβ-Jβ-Cβ). mRNA transcription splices out any intervening sequence and allows translation of the full length protein for the TCR β-chain.
The rearrangement of the alpha (α) chain of the TCR follows β chain rearrangement, and resembles V-to-J rearrangement described for Ig light chains (see above). The assembly of the β- and α- chains results in formation of the αβ-TCR that is expressed on a majority of T cells.
Mechanism
[edit]Key enzymes and components
[edit]The process of V(D)J recombination is mediated by VDJ recombinase, which is a diverse collection of enzymes. The key enzymes involved are recombination activating genes 1 and 2 (RAG), terminal deoxynucleotidyl transferase (TdT), and Artemis nuclease, a member of the ubiquitous non-homologous end joining (NHEJ) pathway for DNA repair.[4] Several other enzymes are known to be involved in the process and include DNA-dependent protein kinase (DNA-PK), X-ray repair cross-complementing protein 4 (XRCC4), DNA ligase IV, non-homologous end-joining factor 1 (NHEJ1; also known as Cernunnos or XRCC4-like factor [XLF]), the recently discovered Paralog of XRCC4 and XLF (PAXX), and DNA polymerases λ and μ.[5] Some enzymes involved are specific to lymphocytes (e.g., RAG, TdT), while others are found in other cell types and even ubiquitously (e.g., NHEJ components).
To maintain the specificity of recombination, V(D)J recombinase recognizes and binds to recombination signal sequences (RSSs) flanking the variable (V), diversity (D), and joining (J) genes segments. RSSs are composed of three elements: a heptamer of seven conserved nucleotides, a spacer region of 12 or 23 basepairs in length, and a nonamer of nine conserved nucleotides. While the majority of RSSs vary in sequence, the consensus heptamer and nonamer sequences are CACAGTG and ACAAAAACC, respectively; and although the sequence of the spacer region is poorly conserved, the length is highly conserved.[6][7] The length of the spacer region corresponds to approximately one (12 basepairs) or two turns (23 basepairs) of the DNA helix. Following what is known as the 12/23 Rule, gene segments to be recombined are usually adjacent to RSSs of different spacer lengths (i.e., one has a "12RSS" and one has a "23RSS").[8] This is an important feature in the regulation of V(D)J recombination.[9]
Process
[edit]V(D)J recombination begins when V(D)J recombinase (through the activity of RAG1) binds a RSS flanking a coding gene segment (V, D, or J) and creates a single-strand nick in the DNA between the first base of the RSS (just before the heptamer) and the coding segment. This is essentially energetically neutral (no need for ATP hydrolysis) and results in the formation of a free 3' hydroxyl group and a 5' phosphate group on the same strand. The reactive hydroxyl group is positioned by the recombinase to attack the phosphodiester bond of opposite strand, forming two DNA ends: a hairpin (stem-loop) on the coding segment and a blunt end on the signal segment.[10] The current model is that DNA nicking and hairpin formation occurs on both strands simultaneously (or nearly so) in a complex known as a recombination center.[11][12][13][14]
The blunt signal ends are flush ligated together to form a circular piece of DNA containing all of the intervening sequences between the coding segments known as a signal joint (although circular in nature, this is not to be confused with a plasmid). While originally thought to be lost during successive cell divisions, there is evidence that signal joints may re-enter the genome and lead to pathologies by activating oncogenes or interrupting tumor suppressor gene function(s)[Ref].
The coding ends are processed further prior to their ligation by several events that ultimately lead to junctional diversity.[15] Processing begins when DNA-PK binds to each broken DNA end and recruits several other proteins including Artemis, XRCC4, DNA ligase IV, Cernunnos, and several DNA polymerases.[16] DNA-PK forms a complex that leads to its autophosphorylation, resulting in activation of Artemis. The coding end hairpins are opened by the activity of Artemis.[17] If they are opened at the center, a blunt DNA end will result; however in many cases, the opening is "off-center" and results in extra bases remaining on one strand (an overhang). These are known as palindromic (P) nucleotides due to the palindromic nature of the sequence produced when DNA repair enzymes resolve the overhang.[18] The process of hairpin opening by Artemis is a crucial step of V(D)J recombination and is defective in the severe combined immunodeficiency (scid) mouse model.
Next, XRCC4, Cernunnos, and DNA-PK align the DNA ends and recruit terminal deoxynucleotidyl transferase (TdT), a template-independent DNA polymerase that adds non-templated (N) nucleotides to the coding end. The addition is mostly random, but TdT does exhibit a preference for G/C nucleotides.[19] As with all known DNA polymerases, the TdT adds nucleotides to one strand in a 5' to 3' direction.[20]
Lastly, exonucleases can remove bases from the coding ends (including any P or N nucleotides that may have formed). DNA polymerases λ and μ then insert additional nucleotides as needed to make the two ends compatible for joining. This is a stochastic process, therefore any combination of the addition of P and N nucleotides and exonucleolytic removal can occur (or none at all). Finally, the processed coding ends are ligated together by DNA ligase IV.[21]
All of these processing events result in a paratope that is highly variable, even when the same gene segments are recombined. V(D)J recombination allows for the generation of immunoglobulins and T cell receptors to antigens that neither the organism nor its ancestor(s) need to have previously encountered, allowing for an adaptive immune response to novel pathogens that develop or to those that frequently change (e.g., seasonal influenza). However, a major caveat to this process is that the DNA sequence must remain in-frame in order to maintain the correct amino acid sequence in the final protein product. If the resulting sequence is out-of-frame, the development of the cell will be arrested, and the cell will not survive to maturity. V(D)J recombination is therefore a very costly process that must be (and is) strictly regulated and controlled.
See also
[edit]- B cell receptor
- T cell receptor
- Basel Institute for Immunology
- Charles M. Steinberg
- NKT cell
- Recombination-activating gene
References
[edit]- ^ "The Nobel Prize in Physiology or Medicine 1987". nobelprize.org. Archived from the original on 13 February 2021. Retrieved 26 December 2014.
- ^ Li A, Rue M, Zhou J, et al. (June 2004). "Utilization of Ig heavy chain variable, diversity, and joining gene segments in children with B-lineage acute lymphoblastic leukemia: implications for the mechanisms of VDJ recombination and for pathogenesis". Blood. 103 (12): 4602–9. doi:10.1182/blood-2003-11-3857. PMID 15010366.
- ^ a b Abbas, Abul K. (2018). "Lymphocyte Development and Antigen Receptor Gene Rearrangement". Cellular and Molecular Immunology (9th ed.). Philadelphia, PA: Elsevier. ISBN 978-0-323-47978-3.
- ^ Ma, Yunmei; Lu, Haihui; Schwarz, Klaus; Lieber, Michael (September 2005). "Repair of Double-Strand DNA Breaks by the Human Nonhomologous DNA End Joining Pathway: the Iterative Processing Model". Cell Cycle. 4 (9): 1193–1200. doi:10.4161/cc.4.9.1977. PMID 16082219.
- ^ Malu, Shruti; Malshetty, Vidyasagar; Francis, Dailia; Cortes, Patricia (2012). "Role of non-homologous end joining in V(D)J recombination". Immunologic Research. 54 (1–3): 233–246. doi:10.1007/s12026-012-8329-z. PMID 22569912. S2CID 45771818.
- ^ Ramsden, Dale; Baetz, Kristin; Wu, Gillian (1994). "Conservation of Sequence in Recombination Signal Sequence Spacers". Nucleic Acids Research. 22 (10): 1785–1796. doi:10.1093/nar/22.10.1785. PMC 308075. PMID 8208601.
- ^ Cowell, Lindsay; Davila, Marco; Ramsden, Dale; Kelsoe, Garnett (2004). "Computational tools for understanding sequence variability in recombination signals". Immunological Reviews. 200: 57–69. doi:10.1111/j.0105-2896.2004.00171.x. PMID 15242396. S2CID 40771963.
- ^ van Gent, Dik; Ramsden, Dale; Gellert, Martin (1996). "The RAG1 and RAG2 Proteins Establish the 12/23 Rule in V(D)J Recombination". Cell. 85 (1): 107–13. doi:10.1016/s0092-8674(00)81086-7. PMID 8620529.
- ^ Hiom, Kevin; Gellert, Martin (1998). "Assembly of a 12/23 Paired Signal Complex: a Critical Control Point in V(D)J Recombination". Molecular Cell. 1 (7): 1011–1019. doi:10.1016/s1097-2765(00)80101-x. PMID 9651584.
- ^ Schatz, David; Swanson, Patrick (2011). "V(D)J Recombination: Mechanisms of Initiation". Annual Review of Genetics. 45: 167–202. doi:10.1146/annurev-genet-110410-132552. PMID 21854230.
- ^ Schatz, David; Ji, Yanhong (2011). "Recombination Centres and the Orchestration of V(D)J Recombination". Nature Reviews Immunology. 11 (4): 251–263. doi:10.1038/nri2941. PMID 21394103. S2CID 33489235.
- ^ Curry, John; Geier, Jamie; Schlissel, Mark (2005). "Single-Strand Recombination Signal Sequence Nicks in vivo: Evidence for a Capture Model of Synapsis". Nature Immunology. 6 (12): 1272–1279. doi:10.1038/ni1270. PMID 16286921. S2CID 10975251.
- ^ Agrawal, Alka; Schatz, David (1997). "RAG1 and RAG2 Form a Stable Postcleavage Synaptic Complex with DNA Containing Signal Ends in V(D)J Recombination". Cell. 89 (1): 43–53. doi:10.1016/s0092-8674(00)80181-6. PMID 9094713.
- ^ Fugmann, Sebastian; Lee, AIfred; Shockett, Penny; Villey, Isabelle; Schatz, David (2000). "The RAG Proteins and V(D)J Recombination: Complexes, Ends, and Transposition". Annual Review of Immunology. 18: 495–527. doi:10.1146/annurev.immunol.18.1.495. PMID 10837067.
- ^ Lewis, Susanna (1994). "The Mechanism of V(D)J Joining: Lessons from Molecular, Immunological, and Comparative Analyses". Advances in Immunology Volume 56. Vol. 56. pp. 27–150. doi:10.1016/s0065-2776(08)60450-2. ISBN 9780120224562. PMID 8073949.
- ^ Helmink, Beth; Sleckman, Barry (2012). "The response to and repair of RAG-mediated DNA double-strand breaks". Annual Review of Immunology. 30: 175–202. doi:10.1146/annurev-immunol-030409-101320. PMC 4038028. PMID 22224778.
- ^ Ma, Yunmei; Schwarz, Klaus; Lieber, Michael (2005). "The Artemis:DNA-PKcs Endonuclease Cleaves DNA Loops, Flaps, and Gaps". DNA Repair. 4 (7): 845–851. doi:10.1016/j.dnarep.2005.04.013. PMID 15936993.
- ^ Lu, Haihui; Schwarz, Klaus; Lieber, Michael (2007). "Extent to Which Hairpin Opening by the Artemis:DNA-PKcs Complex can Contribute to Junctional Diversity in V(D)J Recombination". Nucleic Acids Research. 35 (20): 6917–6923. doi:10.1093/nar/gkm823. PMC 2175297. PMID 17932067.
- ^ Gauss, George; Lieber, Michael (1996). "Mechanistic Constraints on Diversity in Human V(D)J Recombination". Molecular and Cellular Biology. 16 (1): 258–269. doi:10.1128/MCB.16.1.258. PMC 230999. PMID 8524303.
- ^ Benedict, Cindy; Gilfillan, Susan; Thai, To-Ha; Kearney, John (2000). "Terminal Deoxynucleotidyl Transferase and Repertoire Development". Immunological Reviews. 175: 150–157. doi:10.1111/j.1600-065x.2000.imr017518.x. PMID 10933600. S2CID 20634390.
- ^ van Gent, DC; van der Burg, M (10 December 2007). "Non-Homologous End-Joining, a Sticky Affair". Oncogene. 26 (56): 7731–40. doi:10.1038/sj.onc.1210871. PMID 18066085.
Further reading
[edit]- Hartwell LH, Hood L, Goldberg ML, Reynolds AE, Silver LM, Veres RC (2000). Chapter 24, Evolution at the molecular level. In: Genetics. New York: McGraw-Hill. pp. 805–807. ISBN 978-0-07-299587-9.
- V(D)J Recombination. Series: Advances in Experimental Medicine and Biology, Vol. 650 Ferrier, Pierre (Ed.) Landes Bioscience 2009, XII, 199 p. ISBN 978-1-4419-0295-5
