Biomarker
In biomedical contexts, a biomarker, or biological marker, is a measurable indicator of some biological state or condition. Biomarkers are often measured and evaluated using blood, urine, or soft tissues[1] to examine normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.[2] Biomarkers are used in many scientific fields.
Medicine
[edit]Biomarkers used in the medical field, are a part of a relatively new clinical toolset categorized by their clinical applications. The four main classes are molecular, physiologic, histologic and radiographic biomarkers.[3] All four types of biomarkers have a clinical role in narrowing or guiding treatment decisions and follow a sub-categorization of being either predictive, prognostic, or diagnostic.
Predictive
[edit]Predictive molecular, cellular, or imaging biomarkers that pass validation can serve as a method of predicting clinical outcomes. Predictive biomarkers are used to help optimize ideal treatments, and often indicate the likelihood of benefiting from a specific therapy. For example, molecular biomarkers situated at the interface of pathology-specific molecular process architecture and drug mechanism of action promise capturing aspects allowing assessment of an individual treatment response.[4] This offers a dual approach to both seeing trends in retrospective studies and using biomarkers to predict outcomes. For example, in metastatic colorectal cancer predictive biomarkers can serve as a way of evaluating and improving patient survival rates and in the individual case by case scenario, they can serve as a way of sparing patients from needless toxicity that arises from cancer treatment plans.[5]
Common examples of predictive biomarkers are genes such as ER, PR and HER2/neu in breast cancer, BCR-ABL fusion protein in chronic myeloid leukaemia, c-KIT mutations in GIST tumours and EGFR1 mutations in NSCLC.[6]
Diagnostic
[edit]Diagnostic biomarkers that meet a burden of proof can serve a role in narrowing down diagnosis. This can lead to diagnosis that are significantly more specific to individual patients.
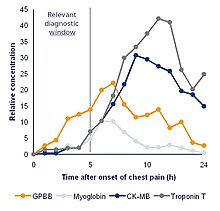
A biomarker can be a traceable substance that is introduced into an organism as a means to examine organ function or other aspects of health.[7]
It can also be a substance whose detection indicates a particular disease state, for example, the presence of an antibody may indicate an infection.[7] More specifically, a biomarker indicates a change in expression or state of a protein that correlates with the risk or progression of a disease, or with the susceptibility of the disease to a given treatment.[7]
One example of a commonly used biomarker in medicine is prostate-specific antigen (PSA). This marker can be measured as a proxy of prostate size with rapid changes potentially indicating cancer. The most extreme case would be to detect mutant proteins as cancer specific biomarkers through selected reaction monitoring (SRM), since mutant proteins can only come from an existing tumor, thus providing ultimately the best specificity for medical purposes.[8]
An example is the traumatic brain injury (TBI) blood-based biomarker test consisted of measuring the levels of neuronal Ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) and Glial fibrillary acidic protein (GFAP) to aid in the diagnosis of the presence of cranial lesion(s) among moderate to mild TBI patients that is(are) otherwise only diagnosable with the use of a CT scan of the head.[9]
Another example is KRAS, an oncogene that encodes a GTPase involved in several signal transduction pathways. Biomarkers for precision oncology are typically utilized in the molecular diagnostics of chronic myeloid leukemia, colon, breast, and lung cancer, and in melanoma.[10]
Digital
[edit]Digital biomarkers are a novel emerging field of biomarkers, mostly collected by smart biosensors.[11] So far, digital biomarkers have been focusing on monitoring vital parameters such as accelerometer data and heartrate[12][13] but also speech.[14] Novel non-invasive, molecular digital biomarkers are increasingly available recorded by e.g. on-skin sweat analysis (internet-enabled Sudorology), which can be seen as next-generation digital biomarkers.[15] Collecting and tracking digital biomarkers have become more easily available with the advancement of smartphones and wearables. In Parkinson's disease (PD), for example, finger tapping a mobile phone via counting apps have been used as a method of (self-)evaluating bradykinesia and effectiveness of medication.[16]
Digital biomarkers are currently being used in conjugation with artificial intelligence (A.I.) in order to recognize symptoms for mild cognitive impairment (MCI).[17] One major current use of digital biomarkers involves keeping track of regular brain activity. Specific neural indicators can be measured by devices to evaluate patients for any neuro abnormalities. The data collected can determine the patients disease probability or condition.[18] While patients carryout everyday tasks (IADL), computers are using machine learning to observe and detect any deviation from normal behavior. These markers are used as signs or indicators of cognitive decline.[17]
Prognostic
[edit]A prognostic biomarker provides information about the patients overall outcome, regardless of any treatment or therapeutic intervention.[6] One example of a prognostic biomarkers in clinical research, is the use of mutated PIK3CA in the study of metastatic breast cancer. As illustrated by the graph, the mutation is prognostic since its presence in the patient endure the same outcome regardless of the treatment method used. Women who had the PIK3CA mutation before treatment, had the lowest average survival rate. The decline in the groups containing the mutant occurred quicker and in a much steeper decline. The independent nature of the prognostic factor allows researcher to study the disease or condition in its natural state. This makes it easier to observe these abnormal biological processes and speculate on how to correct them. Prognostic factors are often used in combination with predictive variables in therapeutics studies, to examine how effective different treatments are in curing specific diseases or cancer. As opposed to predictive biomarkers, prognostic do not rely on any explanatory variables, thus allowing for independent examination of the underlying disease or condition.[19]
Nutrition and diet assessment
[edit]Nutritional biomarkers (biochemical markers of intake) are used to estimate dietary intake in nutrition research, in particular nutritional epidemiology, but also in other disciplines such as archaeology where reliable dietary information are required.[20][21] A nutritional biomarker can be any specimen that reflects intake of dietary constituents and is sufficiently specific.[22][23] Many biomarkers are derived from compounds found in foods, such as sugar or phytochemicals, or combinations thereof using a metabolomics.[20][24] Another type of nutritional biomarkers, in particular common in archaeology, are stable isotope ratios.[25]
Research
[edit]Biomarkers for precision medicine are a part of a relatively new behavioral and clinical toolset. In terms of the behavioral toolset, biomarkers are increasingly being used to motivate health behavior change, particularly in diabetes, cardiovascular diseases, and obesity research.[26] Most research to date uses biomarkers that are easily measured, including weight, blood pressure, and glucose; these biomarkers may reflect the impacts of diet, physical activity, and smoking reduction. However, the methods by which feedback from biomarkers are used in intervention research are varied, and their effectiveness remains unclear.[26]
In reference to the clinical toolset, only two predictive biomarkers are implemented clinically in the case of metastatic colorectal cancer.[5] In this case, the lack of data beyond retrospective studies and successful biomarker-driven approaches may be a factor in using biomarker studies due to the attrition of subjects in clinical trials.[27]
The field of biomarker research is also expanding to include a combinatorial approach to identifying biomarkers from multiple sources. Combining biomarkers from various data allows for the possibility of developing panels that evaluate treatment response based on many biomarkers at a single time. One such area of expanding research in multiple-factor biomarkers is mitochondrial DNA sequencing. Mutations in mitochondrial DNA have been shown to correlate to risk, progression, and treatment response of head and neck squamous cell carcinoma.[28] In this example, a relatively low cost sequencing pipeline was shown to be able to detect low frequency mutations within tumor-associated cells. This highlights the general snapshot capability of mitochondrial DNA-based biomarkers in capturing heterogeneity amongst individuals.[28]
Regulatory validation for clinical use
[edit]The Early Detection Research Network (EDRN) compiled a list of seven criteria by which biomarkers can be assessed in order to streamline clinical validation.[29]
Proof of concept
[edit]Previously used to identify the specific characteristics of the biomarker, this step is essential for doing an in situ validation of these benefits. The biologic rationale of a study must be assessed on a small scale before any large scale studies can occur.[29] Many candidates must be tested to select the most relevant ones.[30]
Analytical performances validation
[edit]One of the most important steps, it serves to identify specific characteristics of the candidate biomarker before developing a routine test.[31] Several parameters are considered including:
Protocol standardization
[edit]This optimizes the validated protocol for routine use, including analysis of the critical points by scanning the entire procedure to identify and control the potential risks.
Ethical issues
[edit]In 1997 the National Institute of Health suggested a need for guidelines and legislation development that would regulate the ethical dimensions of biomarker studies.[29]
Ensuring that all of the participants that are included each step of the project (i.e. planning, implementation, and the compilation of the results) are provided with the protection of ethical principles that are put in place prior to beginning the project. These ethical protections should not only protect the participants in the study, but also the non participants, researchers, sponsors, regulators, and all other persons or groups involved in the study.[29] Some ethical protections could include but are not limited to:[29]
- Informed consent of the participant
- Access to participation opportunities independent of race, socio-economic status, gender, sexuality, etc. (within the range allowed by the experimental protocol)
- Scientific integrity
- Confidentiality of data (anonymity)
- Acknowledgement of conflict of interest in terms of funding and sponsorship by given sponsors
- Transparency and recognition of health and legal risks involved in participation
Cell biology
[edit]In cell biology, a biomarker is a molecule that allows the detection and isolation of a particular cell type (for example, the protein Oct-4 is used as a biomarker to identify embryonic stem cells).[33]
In genetics, a biomarker (identified as genetic marker) is a DNA sequence that causes disease or is associated with susceptibility to disease. They can be used to create genetic maps of whatever organism is being studied.
Applications in chemistry, geology and astrobiology
[edit]A biomarker can be any kind of molecule indicating the existence, past or present, of living organisms. In the fields of geology and astrobiology, biomarkers, versus geomarkers, are also known as biosignatures. The term biomarker is also used to describe biological involvement in the generation of petroleum. Biomarkers were used in the geo-chemical investigation of an oil spill in the San Francisco Bay, California in 1988.[34] On April 22–23 around 400,000 gallons of crude oil was accidentally released into the San Joaquin Valley by a refinery and manufacturing complex of the Shell Oil Company. The oil affected many surrounding areas. Samples of the crude oil were collected in the various regions where it had spread and compared to samples that were unreleased in an attempt to distinguish between the spilled oil and the petrogenic background present in the spill area.[34] Mass Spectra was performed to identify biomarkers and cyclic aliphatic hydrocarbons within the samples. Variations in the concentration of constituents of the crude oil samples and sediments were found.[34]
Ecotoxicology
[edit]Biomarkers are being used to identify the effects of water contamination on aquatic organisms. Benthic macro-invertebrates reside in the sediment on the bottoms of streams, which is where many contaminants settle. These organisms have high exposure to the contamination, which makes them good study species when detecting pollutant concentrations and pollution impacts on an ecosystem.[35] There are a variety of biomarkers within an aquatic organism that can be measured, depending on the contaminant or the response in question. There are also a variety of contaminants within water bodies. To analyze the impact of a pollutant on an organism, the biomarker must respond to a specific contaminant within a specific time frame or at a certain concentration.[36] The biomarkers used to detect pollution in aquatic organisms can be enzymatic or non-enzymatic.[37][38]
Rachel Carson, the author of Silent Spring, raised the issue of using organochlorine pesticides and discussed the possible negative effects that said pesticides have on living organisms.[39] Her book raised ethical issues against chemical corporations that were controlling the general reception of the effect of pesticides on the environment, which pioneered the need for ecotoxicological studies. Ecotoxicologial studies could be considered the precursors to biomarker studies.[40] Biomarkers are used to indicate an exposure to or the effect of xenobiotics which are present in the environment and in organisms. The biomarker may be an external substance itself (e.g. asbestos particles or NNK from tobacco), or a variant of the external substance processed by the body (a metabolite) that usually can be quantified.
History
[edit]The widespread use of the term "biomarker" dates back to as early as 1980.[41] The manner in which the environment was monitored and studied near the end of the 1980s was still mainly reliant on the study of chemical substances that were considered dangerous or toxic when found in moderate concentrations in water, sediments, and aquatic organisms.[40] The methods used to identify these chemical compounds were chromatography, spectrophotometry, electrochemistry, and radiochemistry.[40] Although these methods were successful in elucidating the chemical makeup and concentrations present in the environment of the contaminants and the compounds in question, the tests did not provide data that was informative on the impact of a certain pollutant or chemical on a living organism or ecosystem. It was proposed that characterizing biomarkers could create a warning system to check in on the well being of a population or an ecosystem before a pollutant or compound could wreak havoc on the system. Now, due to the development of biomarker studies, biomarkers can be used and applied in the fields of human medicine and in the detection of diseases.[40]
Definition
[edit]The term "biological marker" was introduced in 1950s.[42][43]
- In 1987, biological markers were defined as "indicators signaling events in biological systems or samples" that could be classified into three categories: exposure, effect and susceptibility markers.[44]
- In 1990, McCarthy and Shugart defined biomarkers as, "measurements at the molecular, biochemical, or cellular level in either wild populations from contaminated habitats or in organisms experimentally exposed to pollutants that indicate that the organism has been exposed to toxic chemicals, and the magnitude of the organism's response".[45]
- In 1994, Depledge defined a biomarker as, "a biochemical, cellular, physiological or behavioral change which can be measured in body tissues or fluids or at the level of the whole organism that reveals the exposure at/or the effects of one or more chemical pollutants."[46]
- In 1996, Van Gestel and Van Brummelen attempted to redefine biomarkers to unambiguously differentiate a biomarker from a bioindicator. According to Van Gestel and Van Brummelen, a biomarker by definition should be used only to describe sublethal biochemical changes resulting from individual exposure to xenobiotics.[47]
- In 1998, the National Institutes of Health Biomarkers Definitions Working Group defined a biomarker as "a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention."[48][2]
- In 2000, De Lafontaine defined the term biomarker as a "biochemical and/or physiological change(s) in organisms exposed to contaminants, and thus represent initial responses to environmental perturbation and contamination".[49]
Active biomonitoring
[edit]De Kock and Kramer developed the concept of active biomonitoring in 1994. Active biomonitoring is a comparison of the chemical/biological properties of a sample that has been relocated to a new environment that contains different conditions than its original environment.[50]
See also
[edit]References
[edit]- ^ Hirsch MS, Watkins J (May 2020). "A Comprehensive Review of Biomarker Use in the Gynecologic Tract Including Differential Diagnoses and Diagnostic Pitfalls". Advances in Anatomic Pathology. 27 (3): 164–192. doi:10.1097/PAP.0000000000000238. PMID 31149908. S2CID 171094630.
- ^ a b Biomarkers Definitions Working Group (March 2001). "Biomarkers and surrogate endpoints: preferred definitions and conceptual framework". Clinical Pharmacology and Therapeutics (Review). 69 (3): 89–95. doi:10.1067/mcp.2001.113989. PMID 11240971. S2CID 288484. as cited in Siderowf A, Aarsland D, Mollenhauer B, Goldman JG, Ravina B (April 2018). "Biomarkers for cognitive impairment in Lewy body disorders: Status and relevance for clinical trials". Movement Disorders (Review). 33 (4): 528–536. doi:10.1002/mds.27355. PMID 29624752. S2CID 4634411.
- ^ Mayeux R (April 2004). "Biomarkers: Potential uses and limitations". NeuroRx. 1 (2): 182–188. doi:10.1602/neurorx.1.2.182. ISSN 1545-5343. PMC 534923. PMID 15717018.
- ^ Lukas A, Heinzel A, Mayer B (March 11, 2019). "Biomarkers for capturing disease pathology as molecular process hyperstructure". bioRxiv 10.1101/573402.
- ^ a b Ruiz-Bañobre J, Kandimalla R, Goel A (March 28, 2019). "Predictive Biomarkers in Metastatic Colorectal Cancer: A Systematic Review". JCO Precision Oncology. 3 (3): 1–17. doi:10.1200/PO.18.00260. PMC 7446314. PMID 32914007.
- ^ a b Oldenhuis CN, Oosting SF, Gietema JA, de Vries EG (May 2008). "Prognostic versus predictive value of biomarkers in oncology". European Journal of Cancer. 44 (7): 946–953. doi:10.1016/j.ejca.2008.03.006. PMID 18396036.
- ^ a b c "NCI Dictionary of Cancer Terms". National Cancer Institute. February 2, 2011. Retrieved February 17, 2020.
- ^ Wang Q, Chaerkady R, Wu J, Hwang HJ, Papadopoulos N, Kopelovich L, et al. (February 2011). "Mutant proteins as cancer-specific biomarkers". Proceedings of the National Academy of Sciences of the United States of America. 108 (6): 2444–2449. Bibcode:2011PNAS..108.2444W. doi:10.1073/pnas.1019203108. PMC 3038743. PMID 21248225.
- ^ Wang KK, Kobeissy F, Shakkour Z, Tyndall J (January 19, 2021). "Thorough overview of ubiquitin C-terminal hydrolase-L1 and glial fibrillary acidic protein as tandem biomarkers cleared by US Food and Drug Administration for the evaluation of intracranial injuries among patients with traumatic brain injury". Acute Med. Surg. 19 (1): e622. doi:10.1002/ams2.622. PMC 7814989. PMID 33510896.
- ^ Nalejska E, Mączyńska E, Lewandowska MA (June 2014). "Prognostic and predictive biomarkers: tools in personalized oncology". Molecular Diagnosis & Therapy. 18 (3): 273–284. doi:10.1007/s40291-013-0077-9. PMC 4031398. PMID 24385403.
- ^ Babrak LM, Menetski J, Rebhan M, Nisato G, Zinggeler M, Brasier N, et al. (2019). "Traditional and Digital Biomarkers: Two Worlds Apart?". Digital Biomarkers. 3 (2): 92–102. doi:10.1159/000502000. PMC 7015353. PMID 32095769.
- ^ Rovini E, Maremmani C, Cavallo F (2017). "How Wearable Sensors Can Support Parkinson's Disease Diagnosis and Treatment: A Systematic Review". Frontiers in Neuroscience. 11: 555. doi:10.3389/fnins.2017.00555. PMC 5635326. PMID 29056899.
- ^ Brasier N, Raichle CJ, Dörr M, Becke A, Nohturfft V, Weber S, et al. (January 2019). "Detection of atrial fibrillation with a smartphone camera: first prospective, international, two-centre, clinical validation study (DETECT AF PRO)". Europace. 21 (1): 41–47. doi:10.1093/europace/euy176. PMC 6321964. PMID 30085018.
- ^ Low DM, Bentley KH, Ghosh SS (February 2020). "Automated assessment of psychiatric disorders using speech: A systematic review". Laryngoscope Investigative Otolaryngology. 5 (1): 96–116. doi:10.1002/lio2.354. PMC 7042657. PMID 32128436.
- ^ Brasier N, Eckstein J (2019). "Sweat as a Source of Next-Generation Digital Biomarkers". Digital Biomarkers. 3 (3): 155–165. doi:10.1159/000504387. PMC 7011725. PMID 32095774.
- ^ Picillo M, Vincos GB, Kern DS, Fox SH, Lang AE, Fasano A (2016). "Learning More from Finger Tapping in Parkinson's Disease: Up and Down from Dyskinesia to Bradykinesia". Movement Disorders Clinical Practice. 3 (2): 184–187. doi:10.1002/mdc3.12246. ISSN 2330-1619. PMC 6353362. PMID 30713911.
- ^ a b Cavedoni S, Chirico A, Pedroli E, Cipresso P, Riva G (2020). "Digital Biomarkers for the Early Detection of Mild Cognitive Impairment: Artificial Intelligence Meets Virtual Reality". Frontiers in Human Neuroscience. 14: 245. doi:10.3389/fnhum.2020.00245. PMC 7396670. PMID 32848660.
- ^ Insel TR (2019). Baumeister H, Montag C (eds.). Digital phenotyping and mobile sensing : new developments in psychoinformatics. Studies in Neuroscience, Psychology and Behavioral Economics (SNPBE). Cham, Switzerland: Springer Cham. p. 14. ISBN 978-3-030-31620-4. OCLC 1126541518.
- ^ Ballman KV (November 2015). "Biomarker: Predictive or Prognostic?". Journal of Clinical Oncology. 33 (33): 3968–3971. doi:10.1200/JCO.2015.63.3651. PMID 26392104.
- ^ a b Jenab M, Slimani N, Bictash M, Ferrari P, Bingham SA (June 2009). "Biomarkers in nutritional epidemiology: applications, needs and new horizons". Human Genetics. 125 (5–6): 507–525. doi:10.1007/s00439-009-0662-5. PMID 19357868. S2CID 24678467.
- ^ Makarewicz CA, Sealy J (April 1, 2015). "Dietary reconstruction, mobility, and the analysis of ancient skeletal tissues: Expanding the prospects of stable isotope research in archaeology". Journal of Archaeological Science. 56: 146–158. Bibcode:2015JArSc..56..146M. doi:10.1016/j.jas.2015.02.035.
- ^ Kaaks R, Riboli E, Sinha R (1997). "Biochemical markers of dietary intake". IARC Scientific Publications (142): 103–126. PMID 9354915.
- ^ Potischman N, Freudenheim JL (March 2003). "Biomarkers of nutritional exposure and nutritional status: an overview". The Journal of Nutrition. 133 (3): 873S–874S. doi:10.1093/jn/133.3.873S. PMID 12612172.
- ^ Tasevska N (July 2015). "Urinary Sugars--A Biomarker of Total Sugars Intake". Nutrients. 7 (7): 5816–5833. doi:10.3390/nu7075255. PMC 4517032. PMID 26184307.
- ^ O'Brien DM (July 17, 2015). "Stable Isotope Ratios as Biomarkers of Diet for Health Research". Annual Review of Nutrition. 35 (1): 565–594. doi:10.1146/annurev-nutr-071714-034511. PMC 4791982. PMID 26048703.
- ^ a b Richardson KM, Jospe MR, Saleh AA, Clarke TN, Bedoya AR, Behrens N, et al. (September 25, 2023). "Use of Biological Feedback as a Health Behavior Change Technique in Adults: Scoping Review". Journal of Medical Internet Research. 25: e44359. doi:10.2196/44359. ISSN 1438-8871. PMC 10562972. PMID 37747766.
- ^ Leon AC, Mallinckrodt CH, Chuang-Stein C, Archibald DG, Archer GE, Chartier K (2006). "Attrition in Randomized Controlled Clinical Trials: Methodological Issues in Psychopharmacology". Biological Psychiatry. 59 (11): 1001–1005. doi:10.1016/j.biopsych.2005.10.020. ISSN 0006-3223. PMID 16503329.
- ^ a b Schubert AD, Channah Broner E, Agrawal N, London N, Pearson A, Gupta A, et al. (February 2020). "Somatic mitochondrial mutation discovery using ultra-deep sequencing of the mitochondrial genome reveals spatial tumor heterogeneity in head and neck squamous cell carcinoma". Cancer Letters. 471: 49–60. doi:10.1016/j.canlet.2019.12.006. PMC 6980748. PMID 31830557.
- ^ a b c d e f The early detection research network :translational research to identify early cancer and cancer risk : initial report. Washington, D.C.: National Cancer Institute, Division of Cancer Prevention. 2000. hdl:2027/mdp.39015049497442.
- ^ "Proof-of-concept study of biomarker development in mice provides a roadmap for a similar approach in humans". Fred Hutchinson Cancer Center. July 19, 2011. Retrieved May 13, 2015.
- ^ Aizpurua-Olaizola O, Toraño JS, Falcon-Perez JM, Williams C, Reichardt N, Boons GJ (March 2018). "Mass spectrometry for glycan biomarker discovery". Trends in Analytical Chemistry. 100: 7–14. doi:10.1016/j.trac.2017.12.015. hdl:1874/364403. Archived from the original on April 1, 2024 – via Utrecht University Repository.
- ^ "Environmental health perspectives. 105/SUPP.4". Environmental Health Perspectives. NIH publication. Retrieved February 17, 2020.
- ^ Biomarkers for Psychiatric Disorders. Publisher: Springer U.S. doi:10.1007/978-0-387-79251-4 Copyright: 2009 ISBN 978-0-387-79250-7 (Print) 978-0-387-79251-4 (Online)
- ^ a b c Hostettler FD (1991). Geochemical investigation of an oil spill in San Francisco Bay, California. Open-file report ;91-130. Menlo Park, CA: US Department of the Interior, Geological Survey. hdl:2027/uc1.31210025025220.
- ^ Wang F, Goulet RR, Chapman PM (December 2004). "Testing sediment biological effects with the freshwater amphipod Hyalella azteca: the gap between laboratory and nature". Chemosphere. 57 (11): 1713–1724. Bibcode:2004Chmsp..57.1713W. doi:10.1016/j.chemosphere.2004.07.050. PMID 15519418.
- ^ Rodrigues C, Guimarães L, Vieira N (August 2019). "Combining biomarker and community approaches using benthic macroinvertebrates can improve the assessment of the ecological status of rivers". Hydrobiologia. 839 (1): 1–24. doi:10.1007/s10750-019-03991-7. ISSN 0018-8158. S2CID 186207664.
- ^ "Online Submission and Review for Science of the Total Environment". Science of the Total Environment. 329 (1): 1. August 15, 2004. Bibcode:2004ScTEn.329....1.. doi:10.1016/j.scitotenv.2004.06.001. ISSN 0048-9697.
- ^ Freitas R, Almeida Â, Pires A, Velez C, Calisto V, Schneider RJ, et al. (November 15, 2015). "The effects of carbamazepine on macroinvertebrate species: Comparing bivalves and polychaetes biochemical responses". Water Research. 85: 137–147. Bibcode:2015WatRe..85..137F. doi:10.1016/j.watres.2015.08.003. ISSN 0043-1354. PMID 26312440.
- ^ Carson R (2000). Silent spring. Penguin Books. ISBN 978-0-14-118494-4. OCLC 934630161.
- ^ a b c d Amiard-Triquet C, Amiard JC, Rainbow PS, eds. (April 19, 2016). Ecological Biomarkers. CRC Press. doi:10.1201/b13036. ISBN 978-0-429-11149-5.
- ^ Aronson JK (May 2005). "Biomarkers and surrogate endpoints". British Journal of Clinical Pharmacology. 59 (5): 491–494. doi:10.1111/j.1365-2125.2005.02435.x. PMC 1884846. PMID 15842546.
- ^ Porter KA (August 1957). "Effect of homologous bone marrow injections in x-irradiated rabbits". British Journal of Experimental Pathology. 38 (4): 401–412. PMC 2082598. PMID 13460185.
- ^ Basu PK, Miller I, Ormsby HL (March 1960). "Sex chromatin as a biologic cell marker in the study of the fate of corneal transplants". American Journal of Ophthalmology. 49 (3): 513–515. doi:10.1016/0002-9394(60)91653-6. PMID 13797463.
- ^ "Biological markers in environmental health research. Committee on Biological Markers of the National Research Council". Environmental Health Perspectives. 74: 3–9. October 1987. doi:10.1289/ehp.87743. PMC 1474499. PMID 3691432.
- ^ McCarthy JF, Shugart LR (January 18, 2018). "Biological Markers of Environmental Contamination". Biomarkers of Environmental Contamination. CRC Press. pp. 3–14. doi:10.1201/9781351070263-2. ISBN 978-1-351-07026-3.
- ^ Depledge MH (January 29, 2020). "The Rational Basis for the Use of Biomarkers as Ecotoxicological Tools". Nondestructive Biomarkers in Vertebrates. CRC Press. pp. 271–295. doi:10.1201/9780367813703-20. ISBN 978-0-367-81370-3. S2CID 81058879.
- ^ Van Gestel CA, Van Brummelen TC (August 1996). "Incorporation of the biomarker concept in ecotoxicology calls for a redefinition of terms". Ecotoxicology. 5 (4): 217–225. Bibcode:1996Ecotx...5..217V. doi:10.1007/bf00118992. PMID 24193812. S2CID 195240026.
- ^ Strimbu K, Tavel JA (November 2010). "What are biomarkers?". Current Opinion in HIV and AIDS. 5 (6): 463–466. doi:10.1097/COH.0b013e32833ed177. PMC 3078627. PMID 20978388.
- ^ Gagné F, Blaise C, Costan G, Gagnon P, Chan HM (August 2000). "Biomarkers in zebra mussels (Dreissena polymorpha) for the assessment and monitoring of water quality of the St Lawrence River (Canada)". Aquatic Toxicology. 50 (1–2): 51–71. Bibcode:2000AqTox..50...51D. doi:10.1016/s0166-445x(99)00094-6. PMID 10930650.
- ^ Smolders R, Voets J, Bervoets L, Blust R, Wepener V (July 15, 2005). "Active Biomonitoring (ABM) by Translocation of Bivalve Molluscs". Water Encyclopedia. John Wiley & Sons, Inc. pp. 33–37. doi:10.1002/047147844x.wq16. ISBN 0-471-47844-X.
